Abstract
Purpose
Methylated cell-free DNA in liquid biopsies are promising non-invasive biomarkers for colorectal cancer (CRC). Optimal markers would have high sensitivity and specificity for early detection of CRC and could be detected in more than one type of material from the patient. We systematically reviewed the literature on DNA methylation markers of colorectal cancer, detected in more than one type of material, regarding their potential as contributors to a panel for screening and follow-up of CRC.
Methods
The databases MEDLINE, Web of Science, and Embase were systematically searched. Data extraction and review was performed by two authors independently. Agreement between methylation status in tissue and other materials (blood/stool/urine) was analyzed using the McNemar test and Cohen’s kappa.
Results
From the 51 included studies, we identified seven single markers with sensitivity ≥ 75% and specificity ≥ 90% for CRC. We also identified one promising plasma panel and two stool panels. The correspondence of methylation status was evaluated as very good for four markers, but only marginal for most of the other markers investigated (12 of 21).
Conclusion
The included studies reported only some of the variables and markers of interest and included few patients. Hence, a meta-analysis was not possible at this point. Larger, prospective studies must be designed to study the discordant detection of markers in tissue and liquid biopsies. When reporting their findings, such studies should use a standardized format.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) develops over years or decades through genetic and epigenetic alterations [1]. CRC is the second leading cause of cancer-related death in the Western world [2, 3], and its treatment and follow-up have a massive impact on the quality of life [4, 5]. Measures to increase CRC survival would be to diagnose the disease at an earlier stage, more reliably identify patients with residual disease after treatment, more accurately diagnose recurrence, and more closely monitor the effect of oncologic treatment. Sensitive and reliable biomarkers in blood, stool, or urine would be ideal for this purpose. Hence, much effort has been put into the identification of new and improved biomarkers for early detection and follow-up of CRC [6, 7]. Cell-free DNA (cfDNA) methylation markers, especially in blood and stool, are considered promising biomarkers, with remarkably high sensitivity and specificity for CRC [8, 9].
Methylation of cytosine to form 5-methylcytosine at CpG dinucleotides is a widespread and normal epigenetic modification of the DNA in humans. Increased CpG methylation in promoter regions of genes, especially at CpG-rich sequences termed CpG islands, is associated with transcription repression [10]. Increased methylation in classical tumor suppressor genes, genes regulating mitosis, and DNA repair is considered an early event in CRC tumorigenesis [1, 11]. Many individual markers have been investigated to date [11,12,13,14,15,16]. However, it is considered that the most useful tool for detection and follow-up of CRC would be a panel of cell-free DNA markers with high sensitivity and specificity for early detection of the disease, with prognostic value, with the possibility to detect residual disease, and recurrence, as well as ability to change as a result of oncologic treatment [17]. Detection even at the adenoma level would be preferable [18]. Also, optimal markers would be possible to detect both in the blood (b), stool (s), urine (u), and tumor tissue (t) throughout the course of the disease [19].
Much of the research on specific methylation markers of CRC has focused on tumor-derived DNA. Lately, single markers and panels of markers have also been tested in tumor remote media such as blood, stool, and urine and have gained much attention as potential liquid biopsies (non-invasive cancer biomarkers) [20, 21]. Somewhat challenging, detection of such markers in blood/urine/stool has not been concordant with detection in CRC tissue [22,23,24]. This inconsistency between liquid biopsies and tissue biopsies has been pointed out as one of the hurdles that need to be addressed before liquid biopsies can be taken into routine clinical use [25]. To overcome this issue, some studies have analyzed methylation markers in several different materials from the same individual. The present review focuses on analyses performed in more than one type of material from the same individual. The objective is to identify cell-free DNA methylation markers of colorectal cancer detected in more than one type of material from the same patient and systematically review their potential as contributors to a panel for screening and follow-up of CRC.
Methods
The search strategy, study inclusion and exclusion criteria, data extraction, study quality assessment, and data analysis were performed based on the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA), the variant for diagnostic test accuracy (PRISMA-DTA), and the quality assessment for diagnostic accuracy studies (QUADAS) [26,27,28,29].
Search strategy, study inclusion, and exclusion
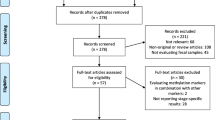
A systematic literature search was performed in MEDLINE, Web of Science, and Embase databases using the following search strategy: (1) «hypermethylation» or «methylation» or «hypermetylation» or «metylation» or «CpG islands», (2) «biomarker» or «liquid biopsy» or «non-invasive» or «ctDNA» or «cfDNA», (3) «colorectal cancer» or «colorectal neoplasm» or «adenoma», (4) («tumour» or «tumor» or «biopsy») or («blood» or «serum» or «plasma» or «blood analysis») or stool or urine, (5) «PCR» or «microarray» or «sequencing», (6) #1 AND #2 AND #3 AND #4 AND #5, (7) «animal» or «cell line», (8) #6 NOT #7. Studies were included if they described one or more DNA methylation markers in more than one type of material from the same individual (blood, stool, urine, or tumor samples) and correlated their finding with CRC. Six recent reviews were hand searched for publications not identified by the systematic search [11,12,13,14,15,16]. Reviews, case reports, and duplicates including conference abstracts later published as full text, studies on animal models or cell lines, as well as papers written in languages other than English were excluded. Study inclusion and exclusion criteria are shown in the PRISMA flowchart (Fig. 1).
Data extraction
Data were extracted in a pre-defined form including the type of specimen, sensitivity for CRC (n/N, %), sensitivity for adenoma (n/N, %), specificity (n/N, %), and method for methylation analysis. Data extraction and review was performed by two authors independently.
Statistical analysis
Agreement between methylation status in tissue and other materials (blood/stool/urine) was analyzed using the McNemar test and Cohen’s kappa. If the p value of the McNemar test was insignificant (p ≥ 0.05) and the p value of the Kappa statistic was significant (p < 0.05), the methylation status was considered as being in agreement. The criteria for the strength of agreement were as follows: K < 0.2 poor, K 0.21–0.40 fair, K 0.41–0.60 moderate, K 0.61–0.80 good, K 0.81–1.00 very good [30]. All statistics were performed using SPSS 25 for windows (SPSS Inc., Chicago, IL, USA)
Results
The systematic literature search identified 728 publications of potential relevance and 85 publications were added by hand search. The abstracts of these 813 publications were reviewed and 381 publications were excluded. Among the remaining, 283 publications considered methylation only in tissue, 51 publications considered methylation only in the blood, 25 publications considered methylation only in stool, and three publications considered methylation only in the bowel lavage (Fig. 1). No publications considered urine only. Methylation in more than one type of material was analyzed in 70 publications, among which only 51 publications analyzed methylation in more than one type of material from the same individual.
The 51 included studies demonstrate that some of the well-known cell-free DNA methylation markers of colorectal cancer can be detected in more than one type of material from the same patient. The extracted data are presented in full as supplementary material (Supplementary Table 1 and 2). Single markers with sensitivity for CRC of 75% or above in tissue and at least one other material were BMP3b, EFHD1b, ITGA4s, NDRG4s, OSMRb PPP1R3Cb, SEPT9b, SFRP1b, SFRP2s, SPG20b,s, TFPI2s, and VIMu (Table 1). Among these, ITGA4, SEPT9, SFRP1, SFRP2, SPG20, TFPI2, and VIM had a specificity of 90% or above (Table 1).
The most promising markers tested as panels were the plasma panel APC/MGMT/RASSF2A/Wif-1 (sensitivity 87%, specificity 92%) [45], the stool panels BMP3/NDRG4/VIM/TFPI2/mutant KRAS/ACTB (β-actin; used as reference gene for normalization purposes)/Hb (sensitivity 87%, specificity 93%) [46], and RARB2/p16/MGMT/APC (sensitivity 75%, specificity 100%) [47] (Table 2).
When investigating the case-by-case relationship for single methylation markers analyzed in more than one type of material from the same patient, we found a very good agreement between tissue and other materials for four markers (CDH4 in the blood, ERCC1 in the blood, p16INK4a in the blood, and SPG20 in stool) (Table 3). Most of the markers (12 of 21) showed marginal reproducibility (k < 0.4) between methylation status in tissue and other materials (Table 3).
Discussion
The present systematic review identified 51 publications analyzing cell-free DNA methylation markers in more than one type of material from the same individual. The markers analyzed in these studies are well known from previous studies in tissue, blood, and stool [11,12,13,14,15,16]. We will here discuss the potential role of each of the identified markers, as contributors to a panel for screening and follow-up of CRC (i.e., sensitivity and specificity for early detection, prognostic value, detection of residual disease and recurrence, ability to reflect ongoing oncologic treatment, and detectability in more than one type of material, i.e., blood (b), stool (s), urine (u), and tumor tissue (t)).
Bone morphogenetic protein 3 (BMP3) is a member of the transforming growth factor-beta (TGFβ) superfamily of cytokines, binding to cell-surface receptors, activating a cascade of cell signaling, ultimately regulating the transcription of SMAD4 target genes to achieve growth suppression [60]. Downregulation of the BMP3 tumor suppressor gene is an early and frequent event in colorectal cancer [60]. In the present systematic review, methylation of BMP3 was identified in 75% of plasma samples and 82% of tissue samples from patients with colorectal cancer [31]. However, methylation of BMP3 was also detected in 11 of the 37 plasma samples from healthy controls referred to colonoscopy with no evidence of CRC or other cancer [31]. Tissue from healthy controls was not analyzed. BMP3 has also been analyzed in other studies where the sensitivity and specificity for CRC in tissue were 57% and 93%, respectively [61], where the sensitivity and specificity for CRC in plasma varied from 29 to 40% and 89 to 94%, respectively [62, 63], and where methylation in stool DNA varied from 40 to 100% in CRC and 33 to 70% in advanced adenoma samples [64, 65]. BMP3 is one of two methylation markers (the other is NDRG4) included in the commercially available fecal Cologuard® test, approved by the FDA in 2014 as a colon cancer screening test [46, 66].
EFHD1 (EF-hand domain family member D1) encodes a calcium-binding protein involved in mitosis, synaptic transmission, and cytoskeletal rearrangement [67]. In the present systematic review, methylation of EFHD1 had 79% sensitivity and 22% specificity for CRC in plasma [32]. When combined with the analysis of PPP1R3C methylation, sensitivity was 53% and specificity reached 96% [32]. No patients with adenoma were included in this study. We have not found other studies on EFHD1 and its value as a potential biomarker for CRC is still unclear.
The ITGA4 gene encodes a membrane protein (integrin alpha 4) and is considered a risk marker for inflammation-associated colon cancer [68]. One study identified in the present systematic review found this marker to have 80% sensitivity and 100% specificity for CRC in stool, and methylated ITGA4 was found in 89% of CRC tissue samples and 88% of adenoma tissue samples [33]. In a stool only study, the sensitivity for adenoma was 29% and the specificity was 69% [69]. Another stool only study found 70% sensitivity and 97% specificity for CRC when investigating a panel combining ITGA4, SFRP2, and p16 [70].
The N-Myc downstream-regulated gene 4 (NDRG4) plays a role in cell growth and differentiation and is a putative tumor suppressor shown to be downregulated in colorectal cancer [71]. In the present systematic review, methylation of NDRG4 had 76% sensitivity for CRC in stool and 73% in urine [34, 72]. No patients with adenomas and no healthy controls were included; hence, no sensitivity for adenoma and no specificity were reported [34, 72]. A study on cell-free DNA in plasma found methylation of NDRG4 in only 18 of 193 patients with CRC (sensitivity 9%) but in none of the healthy controls (specificity 100%) [62]. In two sets of samples from different individuals, NDRG4 methylation analysis in stool found the sensitivity for CRC to be 53 to 61% and the specificity to be 93 to 100% [71]. Methylation was detected in 70 to 86% of CRC tissues as compared with 4% in noncancerous colon mucosa [71]. Despite NDRG4 being included in the Cologuard® test, there are only few studies addressing its role as a potential biomarker.
The oncostatin M receptor gene (OSMR) encodes a subunit of both the oncostatin M (OSM) receptor type II and the interleukin-31 receptor and transduce signals with pro- or anti-proliferative functions. OSMR promoter methylation has been identified in tissue and stool samples from patients with CRC. Two of the included studies confirmed that OSMR methylation is a common epigenetic event in colorectal cancer [35, 38], but one study also revealed low concordance (48%) and specificity (33%) when comparing matched plasma and tumor tissue samples [38].
The protein phosphatase 1 regulatory subunit 3C (PPP1R3C) modulates glycogen metabolism, and methylation of the PPP1R3C gene has been proposed to play a critical role in colorectal cancer. One of the studies included in this review found that methylated PPP1R3C had 92% sensitivity for the detection of stage I CRC [32]. However, the sample size of this study was small and its value as a potential biomarker, especially for early-stage CRC cancer, needs to be further investigated.
SEPT9 encodes a GTP-binding protein involved in cell proliferation and migration, cytokinesis, and angiogenesis [1]. Methylation of SEPT9 is the most extensively studied single methylated marker for colorectal cancer. There are two commercialized assays already in clinical use as blood tests. These are ColoVantage® (sensitivity 90%) [73] and Epi proColon 2.0 (sensitivity 66–81% and specificity 96–99%) [74, 75]. However, the sensitivity in the blood for advanced adenomas has been < 10% in several studies [76,77,78]. In the present systematic review, the most well-designed studies generally found a good correlation between the methylation status of SEPT9 in plasma and tissue for CRC patients and also for healthy controls [35, 36, 79, 80]. In patients with adenomas the impression is that even though patients have methylated SEPT9 in tissue, only one-third of them have detectable methylated SEPT9 in plasma [37]. The reason for this discrepancy remains unknown.
The secreted frizzled-related protein 1 and 2 genes (SFRP1 and SFRP2) encode antagonists of the Wnt signaling pathway, acting as tumor suppressors. Altered methylation of these genes has been reported in colorectal cancer tissue [81]. Aberrant promoter methylation of SFRP2 is associated with poor survival of colorectal cancer [39, 82]. Two well-designed studies found the sensitivity of SFRP2 for CRC to be above 85% in stool and ranging from 46 to 62% for adenoma, the specificity was above 90%, and the findings corresponded well with the findings in tissue for the same patients [39, 40]. In studies of only one type of material, Rasmussen et al. found that the sensitivity of SFRP2 for CRC to be 21% for CRC and the specificity was 82% in plasma [62]. In stool only studies, sensitivity for CRC was 63–94%, sensitivity for adenomas was 46–92%, and specificity was 32–100% [83,84,85,86,87]. As mentioned previously, the combination of SFRP2 with other markers, such as VIM, RASSF1A, TMEFF2, MGMT, ITGA4, and p16, has provided promising results [70, 72, 85, 87, 88].
The spastic paraplegia 20 gene (SPG20) encodes Spartin, a multifunctional protein found to be involved in intracellular epidermal growth factor receptor trafficking [89]. In the present systematic review, we identified two studies investigating this biomarker in more than one type of material, although none of these studies included adenomas. Rezvani et al. found methylated SPG20 in 94% of CRC tissues and correspondingly in 81% of the plasma samples from these patients [41]. The specificity in tissue samples was 99% and in plasma samples 97% [41]. Zhang et al. found methylated SPG20 in 85% of CRC tissues and correspondingly in 80% of stool samples from these patients [42]. No tissue samples from healthy controls were investigated, but the specificity in the stool was 100% [42]. In a study carried out in plasma only, the sensitivity of SPG20 was 16% for CRC and the specificity was 88%, whereas the sensitivity for adenomas was not investigated [62].
The tissue factor pathway inhibitor 2 gene (TFPI2) encodes a serine protease inhibitor that decreases the activities of several enzymes, thereby protecting the extracellular matrix from degradation and inhibiting in vitro colony formation and proliferation [68]. Loss of TFPI2 function has been associated with pro-invasiveness and methylation of TFPI2 is considered an independent prognostic factor for CRC as it has been associated with later stages of carcinogenesis and advanced colorectal cancer [68, 90]. TFPI2 has been demonstrated to be completely unmethylated in tissue from healthy controls, whereas methylation frequency increases with progression into inflamed colon tissue or CRC [68]. In the present review, two studies investigating TFPI2 methylation in stool and corresponding tissues found methylation of TFPI2 in CRC tissues in 89% and 99% of the samples, respectively, and the sensitivity in stool was 80% and 76%, respectively [33, 43]. Specificity for CRC in stool was 100% and 93%, respectively [33, 43]. Glöckner et al. also found methylated TFPI2 in three of the tissue samples from healthy controls (6%) and in 55 of 56 adenoma samples (98%) [43]. However, the sensitivity of TFPI2 methylation in adenoma stool samples was only 21% [43]. In a study of plasma, the CRC sensitivity of TFPI2 methylation was only 7%, no adenomas were included, and the specificity was 98% [62]. In a study of stool, the sensitivity for CRC was 68%, the specificity was 100%, whereas the sensitivity for adenoma was 35% [91, 92].
VIM encodes a protein of the cytoskeleton. VIM is considered a biomarker of mesenchymal-derived cells and cancer cells undergoing epithelial-mesenchymal transition (EMT) during invasion and metastasis, and promoter methylation of VIM has been documented in CRC [1]. ColoSure® is a fecal single-marker test used in combination with colonoscopy. The test detects methylated VIM as a marker for CRC (sensitivity38–88%) [1, 93]. In paired samples from patients with CRC and/or adenomas, the share with detectable methylated VIM was 33% in stool, 4% in serum, and 8% in urine and the specificity was 100% in all materials [94]. In one study, where 83% of CRC cases had methylated VIM in tissue samples, 55% also had methylated VIM in stool [95]. In another study, where 44% of CRC cases had methylated VIM in tissue, 40% also had methylated VIM in stool [33]. Gerecke et al. also found that 72% of patients with adenomas had methylated VIM in adenoma tissue [33]. In Song et al.’s study, 85% of CRC cases had methylated VIM in tissue samples and 75% also had methylated VIM in urine [44]. When combining analysis of SFRP2 and VIM, methylation in at least one of these was found in 92% of CRC and 94% of adenoma tissues and correspondingly in 89% and 85% of stool samples [72]. VIM methylation is extensively studied in unpaired samples. The sensitivity for CRC detection by VIM methylation in the blood is 18–59% and the specificity is 63–93% [62, 96, 97]. The sensitivity for CRC in stool is 38–85%, and the sensitivity and specificity for adenoma are 75–90% and 82–100 % respectively [98,99,100,101,102,103].
One plasma panel consisting of the methylation markers APC, MGMT, RASSF2A, and WIF1 was identified by the present systematic review as promising, by reporting 86% sensitivity and 92% specificity for CRC [45]. Methylation in corresponding fresh-frozen tissue samples was found to be 27% for APC (specificity 97%), 39% for MGMT (specificity 96%), 58% for RASSF2A (specificity 100%), and 74% for WIF1 (specificity 98%) [45]. Methylation of APC leads to activation of growth-promoting genes, methylation of MGMT impairs the elimination of DNA alkylation adducts which may lead to mutation, methylation of the tumor suppressor RASSF2 alters the regulation of the cytoskeleton and apoptosis, and methylation of WIF1 results in reduced blocking of the Wnt signaling pathway [45].
We also identified two stool panels as the most promising panels tested in more than one type of material from the same individual. One was the panel consisting of methylated BMP3/NDRG4/VIM/TFPI2/mutant KRAS, β-actin, and Hb-level (and later refined and commercialized as Cologuard®), which in stool detected CRC with 87% sensitivity, adenoma with 82% sensitivity, and demonstrated a specificity of 93% [46, 48, 66]. Corresponding plasma samples were tested for methylated SEPT9 and the reported sensitivity for CRC was 60%, adenoma 14%, and the specificity was 73% [48]. The second stool panel identified was one consisting of RARB2, p16, MGMT, and APC. This panel detected CRC with 75% sensitivity, adenomas with 60% sensitivity, and the specificity was 100% [49]. To our knowledge, no other research groups have reproduced these findings and the findings have not led to a commercialized test.
Our overall impression is that the quality of the included studies was variable. With reference to QUADAS; many studies do not clearly classify the patients with colorectal cancer (some also include patients with adenomas and polyps), many have a sub-optimal or mixed “control group” (self-declared healthy controls or the control group including inflammatory bowel disease, colitis, polyps), many do not report blinding (i.e., it is not known whether the researcher who performed the methylation analysis knew whether the patient was a healthy control or a cancer patient), and many studies fail to report important findings (“not specified” in Supplementary Tables 1 and 2). To determine the strength of each methylated marker, it would be useful to aggregate the findings from single studies into a meta-analysis. Currently, this is not possible due to inconsistency in the reporting. To ensure comparability and future aggregation of findings, we suggest that future studies report their findings according to Table 4.
During the present review, we found that many of the studies claiming to report methylation in so-called paired samples do not analyze corresponding materials from the same individual. In addition, few of the studies reported the absolute numbers needed to calculate sensitivity and sensitivity. For the studies reporting the absolute numbers of methylation status in tissue and corresponding other materials from single individuals, we performed a case-by-case analysis (Table 3). The correspondence of methylation status in different materials from the same individual was promising for CDH4, ERCC1, p16INK4a, and SPG20. However, for most of the methylation markers and in most of the corresponding materials investigated (16 of 27), only a marginal reproducibility of methylation status was found. This inconsistency between “liquid biopsies” and tissue biopsies is confusing and needs to be addressed by analyzing methylation markers in several different materials from the same individual. And, this needs to be done both for patients with CRC, patients with adenoma, and healthy controls.
In the publications reviewed here, we rediscover some of the known technical challenges to the practical and clinical application of methylated cell-free DNA markers [58]. To date, there are still no standards agreed upon for analysis of methylated cell-free DNA and among the included publications, there is considerable variation regarding the methods used to extract and quantify cell-free DNA. The methods for sample processing and analysis also vary a lot, and many of the studies do not address or report upon pre-analytical and analytical issues. A study on optimization of the yield and utility of circulating cell-free DNA showed that time from blood sampling to plasma separation was extremely significant, whereas the temperature of the blood sample before plasma separation (kept on ice or at room temperature) did not affect the amount of cell-free DNA significantly [104]. One report has stated that DNA levels in plasma stored at − 80 °C declines by 30% per year [105]. The cell-free DNA methylation pattern may be affected by demographic and lifestyle characteristics (i.e., age, race, gender, smoking, alcohol consumption), diet (i.e., folate, green tea, phytoestrogen), environmental exposures (i.e., arsenic, cadmium), and disease status [106]. Only a few of the studies take such factors into account when analyzing and reporting their results. The small sample sizes, selective and incomplete reporting, and lack of independent validation of promising biomarkers are factors hampering the implementation of potential biomarkers into clinical use [107]. The use of sensitive technologies, unbiased optimization, and standardization of commonly used methods is utterly important to enable validation of the findings from promising biomarker discovery studies [108].
Conclusion
The identified studies analyzing methylated DNA markers in paired samples from the same individuals generally included few patients, reported only some of the variables of interest and investigated only some of the markers of interest. Hence, a meta-analysis was not possible at this point. Larger, prospective studies need to be designed to overcome the challenge of discordant detection of markers in tissue and liquid biopsies. To improve comparability and to facilitate aggregation of the evidence base regarding possible DNA methylation markers of colorectal cancer, we suggest that this type of study use a standardized format when reporting their findings.
Availability of data and material
Not applicable.
References
El Bairi K, Tariq K, Himri I, Jaafari A, Smaili W, Kandhro AH, Gouri A, Ghazi B (2018) Decoding colorectal cancer epigenomics. Cancer Gene Ther 220:49–76. https://doi.org/10.1016/j.cancergen.2017.11.001
Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, Gavin A, Visser O, Bray F (2018) Cancer incidence and mortality patterns in Europe: estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer (Oxford, England: 1990) 103:356–387. https://doi.org/10.1016/j.ejca.2018.07.005
Siegel RL, Miller KD, Jemal A (2019) Cancer statistics, 2019. CA Cancer J Clin 69(1):7–34. https://doi.org/10.3322/caac.21551
Cabilan CJ, Hines S (2017) The short-term impact of colorectal cancer treatment on physical activity, functional status and quality of life: a systematic review. JBI Database System Rev Implement Rep 15(2):517–566. https://doi.org/10.11124/JBISRIR-2016003282
Anthony T, Jones C, Antoine J, Sivess-Franks S, Turnage R (2001) The effect of treatment for colorectal cancer on long-term health-related quality of life. Ann Surg Oncol 8(1):44–49
Cuyle PJ, Prenen H (2017) Current and future biomarkers in the treatment of colorectal cancer. Acta Clin Belg 72(2):103–115. https://doi.org/10.1080/17843286.2016.1262996
Boussios S, Ozturk MA, Moschetta M, Karathanasi A, Zakynthinakis-Kyriakou N, Katsanos KH, Christodoulou DK, Pavlidis N (2019) The developing story of predictive biomarkers in colorectal cancer. J Pers Med 9(1). https://doi.org/10.3390/jpm9010012
Worm Orntoft MB (2018) Review of blood-based colorectal cancer screening: how far Are circulating cell-free DNA methylation markers from clinical implementation? Clin Colorectal Cancer 17(2):e415–e433. https://doi.org/10.1016/j.clcc.2018.02.012
Gabriel E, Bagaria SP (2018) Assessing the impact of circulating tumor DNA (ctDNA) in patients with colorectal cancer: separating fact from fiction. Front Oncol 8:297. https://doi.org/10.3389/fonc.2018.00297
Zamani M, Hosseini SV, Mokarram P (2018) Epigenetic biomarkers in colorectal cancer: premises and prospects. Biomarkers 23(2):105–114. https://doi.org/10.1080/1354750X.2016.1252961
Goel A, Boland CR (2012) Epigenetics of colorectal cancer. Gastroenterology 143(6):1442–1460 e1441. https://doi.org/10.1053/j.gastro.2012.09.032
Vedeld HM, Goel A, Lind GE (2018) Epigenetic biomarkers in gastrointestinal cancers: The current state and clinical perspectives. Semin Cancer Biol 51:36–49. https://doi.org/10.1016/j.semcancer.2017.12.004
Rasmussen SL, Krarup HB, Sunesen KG, Pedersen IS, Madsen PH, Thorlacius-Ussing O (2016) Hypermethylated DNA as a biomarker for colorectal cancer: a systematic review. Color Dis 18(6):549–561. https://doi.org/10.1111/codi.13336
Okugawa Y, Grady WM, Goel A (2015) Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology 149(5):1204–1225 e1212. https://doi.org/10.1053/j.gastro.2015.07.011
Hashimoto Y, Zumwalt TJ, Goel A (2016) DNA methylation patterns as noninvasive biomarkers and targets of epigenetic therapies in colorectal cancer. Epigenomics 8(5):685–703. https://doi.org/10.2217/epi-2015-0013
Kim MS, Lee J, Sidransky D (2010) DNA methylation markers in colorectal cancer. Cancer Metastasis Rev 29(1):181–206. https://doi.org/10.1007/s10555-010-9207-6
Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N (2017) Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17(4):223–238. https://doi.org/10.1038/nrc.2017.7
Holme O, Loberg M, Kalager M, Bretthauer M, Hernan MA, Aas E, Eide TJ, Skovlund E, Schneede J, Tveit KM, Hoff G (2014) Effect of flexible sigmoidoscopy screening on colorectal cancer incidence and mortality: a randomized clinical trial. JAMA 312(6):606–615. https://doi.org/10.1001/jama.2014.8266
Thomas ML, Marcato P (2018) Epigenetic modifications as biomarkers of tumor development, therapy response, and recurrence across the cancer care continuum. Cancers 10(4). https://doi.org/10.3390/cancers10040101
Zeng H, He B, Yi CQ, Peng JY (2018) Liquid biopsies: DNA methylation analyses in circulating cell-free DNA. J Genet Genomics 45(4):185–192. https://doi.org/10.1016/j.jgg.2018.02.007
Mahasneh A, Al-Shaheri F, Jamal E (2017) Molecular biomarkers for an early diagnosis, effective treatment and prognosis of colorectal cancer: current updates. Exp Mol Pathol 102(3):475–483. https://doi.org/10.1016/j.yexmp.2017.05.005
Symonds EL, Pedersen SK, Murray DH, Jedi M, Byrne SE, Rabbitt P, Baker RT, Bastin D, Young GP (2018) Circulating tumour DNA for monitoring colorectal cancer-a prospective cohort study to assess relationship to tissue methylation, cancer characteristics and surgical resection. Clin Epigenetics 10:63. https://doi.org/10.1186/s13148-018-0500-5
Roperch JP, Incitti R, Forbin S, Bard F, Mansour H, Mesli F, Baumgaertner I, Brunetti F, Sobhani I (2013) Aberrant methylation of NPY, PENK, and WIF1 as a promising marker for blood-based diagnosis of colorectal cancer. BMC Cancer 13:566. https://doi.org/10.1186/1471-2407-13-566
Hellebrekers DM, Lentjes MH, van den Bosch SM, Melotte V, Wouters KA, Daenen KL, Smits KM, Akiyama Y, Yuasa Y, Sanduleanu S, Khalid-de Bakker CA, Jonkers D, Weijenberg MP, Louwagie J, van Criekinge W, Carvalho B, Meijer GA, Baylin SB, Herman JG, de Bruine AP, van Engeland M (2009) GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin Cancer Res 15(12):3990–3997. https://doi.org/10.1158/1078-0432.CCR-09-0055
Jia S, Zhang R, Li Z, Li J (2017) Clinical and biological significance of circulating tumor cells, circulating tumor DNA, and exosomes as biomarkers in colorectal cancer. Oncotarget 8(33):55632–55645. https://doi.org/10.18632/oncotarget.17184
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6(7):e1000097. https://doi.org/10.1371/journal.pmed.1000097
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D (2009) The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med 6(7):e1000100. https://doi.org/10.1371/journal.pmed.1000100
Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J (2003) The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol 3:25. https://doi.org/10.1186/1471-2288-3-25
McGrath TA, Moher D, McInnes MDF (2019) Steps toward more complete reporting of systematic reviews of diagnostic test accuracy: Preferred Reporting Items for Systematic Reviews and Meta-Analyses of Diagnostic Test Accuracy (PRISMA-DTA). Syst Rev 8(1):166. https://doi.org/10.1186/s13643-019-1090-9
Shalaby SM, El-Shal AS, Abdelaziz LA, Abd-Elbary E, Khairy MM (2018) Promoter methylation and expression of DNA repair genes MGMT and ERCC1 in tissue and blood of rectal cancer patients. Gene 644:66–73. https://doi.org/10.1016/j.gene.2017.10.056
Ashoori H, Ghamarchehreh ME, Tavallaei M, Ganji SM, Hosseini M, Zolfaghari M, Ghamarchehreh Z, Vahidian F (2018) Evaluation of the epigenetic biomarker bone morphogenic protein 3 for colorectal cancer diagnosis. J Clin Diagn Res 12(11). https://doi.org/10.7860/JCDR/2018/37101.12260
Takane K, Midorikawa Y, Yagi K, Sakai A, Aburatani H, Takayama T, Kaneda A (2015) Aberrant promoter methylation of PPP1R3C and EFHD1 in plasma of colorectal cancer patients. Cancer Med 3(5):1235–1245. https://doi.org/10.1002/cam4.273
Gerecke C, Lowenstein Y, Fait I, Scholtka B (2012) Comparison of three putative methylation markers for the early detection of colon cancer. J Cancer Res Clin Oncol 1:41–42. https://doi.org/10.1007/s00432-011-1144-4
Xiao W, Zhao H, Dong W, Li Q, Zhu J, Li G, Zhang S, Ye M (2015) Quantitative detection of methylated NDRG4 gene as a candidate biomarker for diagnosis of colorectal cancer. Oncol Lett 9(3):1383–1387. https://doi.org/10.3892/ol.2014.2815
Yuan P, Cheng XJ, Wu XJ, Li L, Zhang LH, Li ZY, Xing XF, Du H, Wang XH, Hu Y, Zhang QY, Deng GR, Wen XZ, Ji JF (2016) OSMR and SEPT9: promising biomarkers for detection of colorectal cancer based on blood-based tests. Transl Cancer Res 5(2):131–139. https://doi.org/10.21037/tcr.2016.03.07
He QO, Chen HY, Bai EQ, Luo YX, Fu RJ, He YS, Jiang J, Wang HQ (2010) Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet Cytogenet 202(1):1–10. https://doi.org/10.1016/j.cancergencyto.2010.05.018
Toth K, Wasserkort R, Sipos F, Kalmaar A, Wichmann B, Leiszter K, Valcz G, Juhasz M, Miheller P, Patai AV, Tulassay Z, Molnar B (2014) Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS One 9 (12). doi:https://doi.org/10.1371/journal.pone.0115415 e115415
Bedin C, Enzo MV, Del Bianco P, Pucciarelli S, Nitti D, Agostini M (2017) Diagnostic and prognostic role of cell-free DNA testing for colorectal cancer patients. Int J Cancer 140(8):1888–1898. https://doi.org/10.1002/ijc.30565
Tang D, Liu J, Wang DR, Yu HF, Li YK, Zhang JQ (2011) Diagnostic and prognostic value of the methylation status of secreted frizzled-related protein 2 in colorectal cancer. Clin Invest Med 34(2):E88–E95
Wang DR, Tang D (2008) Hypermethylated SFRP2 gene in fecal DNA is a high potential biomarker for colorectal cancer noninvasive screening. World J Gastroenterol 14(4):524–531
Rezvani N, Alibakhshi R, Vaisi-Raygani A, Bashiri H, Saidijam M (2017) Detection of SPG20 gene promoter-methylated DNA, as a novel epigenetic biomarker, in plasma for colorectal cancer diagnosis using the MethyLight method. Oncol Lett 13(5):3277–3284. https://doi.org/10.3892/ol.2017.5815
Zhang H, Song YC, Dang CX (2013) Detection of hypermethylated spastic paraplegia-20 in stool samples of patients with colorectal cancer. Int J Med Sci 10(3):230–234. https://doi.org/10.7150/ijms.5278
Glockner SC, Dhir M, Yi JM, McGarvey KE, Van Neste L, Louwagie J, Chan TA, Kleeberger W, de Bruine AP, Smits KM, Khalid-de Bakker CA, Jonkers DM, Stockbrugger RW, Meijer GA, Oort FA, Iacobuzio-Donahue C, Bierau K, Herman JG, Baylin SB, Van Engeland M, Schuebel KE, Ahuja N (2009) Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res 69(11):4691–4699. https://doi.org/10.1158/0008-5472.CAN-08-0142
Song BP, Jain S, Lin SY, Chen Q, Block TM, Song W, Brenner DE, Su YH (2012) Detection of hypermethylated vimentin in urine of patients with colorectal cancer. J Mol Diagn 14(2):112–119. https://doi.org/10.1016/j.jmoldx.2011.12.003
Lee BB, Lee EJ, Jung EH, Chun HK, Chang DK, Song SY, Park J, Kim DH (2009) Aberrant methylation of APC, MGMT, RASSF2A, and Wif-1 genes in plasma as a biomarker for early detection of colorectal cancer. Clin Cancer Res 15(19):6185–6191. https://doi.org/10.1158/1078-0432.Ccr-09-0111
Ahlquist DA, Zou H, Domanico M, Mahoney DW, Yab TC, Taylor WR, Butz ML, Thibodeau SN, Rabeneck L, Paszat LF, Kinzler KW, Vogelstein B, Bjerregaard NC, Laurberg S, Sorensen HT, Berger BM, Lidgard GP (2012) Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology 142(2):248–256; quiz e225-246. https://doi.org/10.1053/j.gastro.2011.10.031
Azuara D, Rodriguez-Moranta F, Soriano-Izquierdo A, Guardiola J, De Oca J, Biondo S, Blanco I, Esteller M, Capella G (2009) Evaluation of stool melting curve analysis of methylated CpG island promoters as an alternative for early noninvasive diagnosis of colorectal tumors. J Clin Oncol 1:e15036
Ahlquist DA, Taylor WR, Mahoney DW, Zou H, Domanico M, Thibodeau SN, Boardman LA, Berger BM, Lidgard GP (2012) The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. Clin Gastroenterol Hepatol 10(3):272–277 e271. https://doi.org/10.1016/j.cgh.2011.10.008
Azuara D, Rodriguez-Moranta F, de Oca J, Soriano-Izquierdo A, Mora J, Guardiola J, Biondo S, Blanco I, Peinado MA, Moreno V, Esteller M, Capella G (2010) Novel methylation panel for the early detection of colorectal tumors in stool DNA. Clin Colorectal Cancer 9(3):168–176. https://doi.org/10.3816/CCC.2010.n.023
Miotto E, Sabbioni S, Veronese A, Calin GA, Gullini S, Liboni A, Gramantieri L, Bolondi L, Ferrazzi E, Gafa R, Lanza G, Negrini M (2004) Frequent aberrant methylation of the CDH4 gene promoter in human colorectal and gastric cancer. Cancer Res 64(22):8156–8159. https://doi.org/10.1158/0008-5472.CAN-04-3000
Nishioka Y, Ueki T, Hokazono K, Nagayoshi K, Tanaka M (2015) Comparative detection of aberrantly methylated DNA in preoperative and postoperative stool from patients with colorectal cancers. Int J Biol Markers 30(1):e81–e87. https://doi.org/10.5301/jbm.5000099
Yamaguchi S, Asao T, Nakamura J, Ide M, Kuwano H (2003) High frequency of DAP-kinase gene promoter methylation in colorectal cancer specimens and its identification in serum. Cancer Lett 194(1):99–105
Philipp AB, Stieber P, Nagel D, Neumann J, Spelsberg F, Jung A, Lamerz R, Herbst A, Kolligs FT (2012) Prognostic role of methylated free circulating DNA in colorectal cancer. Int J Cancer 131(10):2308–2319. https://doi.org/10.1002/ijc.27505
Frattini M, Gallino G, Signoroni S, Balestra D, Lusa L, Battaglia L, Sozzi G, Bertario L, Leo E, Pilotti S, Pierotti MA (2008) Quantitative and qualitative characterization of plasma DNA identifies primary and recurrent colorectal cancer. Cancer Lett 263(2):170–181. https://doi.org/10.1016/j.canlet.2008.03.021
Nakayama G, Kodera Y, Ohashi N, Koike M, Fujiwara M, Nakao A (2011) p16INK4a methylation in serum as a follow-up marker for recurrence of colorectal cancer. Anticancer Res 31(5):1643–1646
Nakayama H, Hibi K, Takase T, Yamazaki T, Kasai Y, Ito K, Akiyama S, Nakao A (2003) Molecular detection of p16 promoter methylation in the serum of recurrent colorectal cancer patients. Int J Cancer 105(4):491–493. https://doi.org/10.1002/ijc.11117
Nakayama H, Hibi K, Taguchi M, Takase T, Yamazaki T, Kasai Y, Ito K, Akiyama S, Nakao A (2002) Molecular detection of p16 promoter methylation in the serum of colorectal cancer patients. Cancer Lett 188(1-2):115–119. https://doi.org/10.1016/s0304-3835(01)00839-4
Danese E, Minicozzi AM, Benati M, Montagnana M, Paviati E, Salvagno GL, Gusella M, Pasini F, Guidi GC, Lippi G (2013) Epigenetic alteration: new insights moving from tissue to plasma - the example of PCDH10 promoter methylation in colorectal cancer. Br J Cancer 109(3):807–813. https://doi.org/10.1038/bjc.2013.351
Wang YC, Yu ZH, Liu C, Xu LZ, Yu W, Lu J, Zhu RM, Li GL, Xia XY, Wei XW, Ji HZ, Lu H, Gao Y, Gao WM, Chen LB (2008) Detection of RASSF1A promoter hypermethylation in serum from gastric and colorectal adenocarcinoma patients. World J Gastroenterol 14(19):3074–3080
Loh K, Chia JA, Greco S, Cozzi SJ, Buttenshaw RL, Bond CE, Simms LA, Pike T, Young JP, Jass JR, Spring KJ, Leggett BA, Whitehall VL (2008) Bone morphogenic protein 3 inactivation is an early and frequent event in colorectal cancer development. Genes Chromosom Cancer 47(6):449–460. https://doi.org/10.1002/gcc.20552
Houshmand M, Abbaszadegan MR, Kerachian MA (2017) Assessment of bone morphogenetic protein 3 methylation in Iranian patients with colorectal cancer. Middle East J Dig Dis 9(3):158–163. https://doi.org/10.15171/mejdd.2017.67
Rasmussen SL, Krarup HB, Sunesen KG, Johansen MB, Stender MT, Pedersen IS, Madsen PH, Thorlacius-Ussing O (2017) Hypermethylated DNA, a circulating biomarker for colorectal cancer detection. PLoS One 12(7):e0180809. https://doi.org/10.1371/journal.pone.0180809
Rokni P, Shariatpanahi AM, Sakhinia E, Kerachian MA (2018) BMP3 promoter hypermethylation in plasma-derived cell-free DNA in colorectal cancer patients. Genes Genomics 40(4):423–428. https://doi.org/10.1007/s13258-017-0644-2
Kisiel JB, Yab TC, Nazer Hussain FT, Taylor WR, Garrity-Park MM, Sandborn WJ, Loftus EV, Wolff BG, Smyrk TC, Itzkowitz SH, Rubin DT, Zou H, Mahoney DW, Ahlquist DA (2013) Stool DNA testing for the detection of colorectal neoplasia in patients with inflammatory bowel disease. Aliment Pharmacol Ther 37(5):546–554. https://doi.org/10.1111/apt.12218
Park SK, Song CS, Yang HJ, Jung YS, Choi KY, Koo DH, Kim KE, Jeong KU, Kim HO, Kim H, Chun HK, Park DI (2016) Field cancerization in sporadic colon cancer. Gut Liver 10(5):773–780. https://doi.org/10.5009/gnl15334
Imperiale TF, Ransohoff DF, Itzkowitz SH (2014) Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 371(2):187–188. https://doi.org/10.1056/NEJMc1405215
Dutting S, Brachs S, Mielenz D (2011) Fraternal twins: Swiprosin-1/EFhd2 and Swiprosin-2/EFhd1, two homologous EF-hand containing calcium binding adaptor proteins with distinct functions. Cell Commun Signal 9:2. https://doi.org/10.1186/1478-811X-9-2
Gerecke C, Scholtka B, Lowenstein Y, Fait I, Gottschalk U, Rogoll D, Melcher R, Kleuser B (2015) Hypermethylation of ITGA4, TFPI2 and VIMENTIN promoters is increased in inflamed colon tissue: putative risk markers for colitis-associated cancer. J Cancer Res Clin Oncol 141(12):2097–2107. https://doi.org/10.1007/s00432-015-1972-8
Ausch C, Kim YH, Tsuchiya KD, Dzieciatkowski S, Washington MK, Paraskeva C, Radich J, Grady WM (2009) Comparative analysis of PCR-based biomarker assay methods for colorectal polyp detection from fecal DNA. Clin Chem 55(8):1559–1563. https://doi.org/10.1373/clinchem.2008.122937
Chang E, Park DI, Kim YJ, Kim BK, Park JH, Kim HJ, Cho YK, Sohn CI, Jeon WK, Kim BI, Kim HD, Kim DH, Kim YH (2010) Detection of colorectal neoplasm using promoter methylation of ITGA4, SFRP2, and p16 in stool samples: a preliminary report in Korean patients. Hepato-gastroenterology 57(101):720–727
Melotte V, Lentjes MH, van den Bosch SM, Hellebrekers DM, de Hoon JP, Wouters KA, Daenen KL, Partouns-Hendriks IE, Stessels F, Louwagie J, Smits KM, Weijenberg MP, Sanduleanu S, Khalid-de Bakker CA, Oort FA, Meijer GA, Jonkers DM, Herman JG, de Bruine AP, van Engeland M (2009) N-Myc downstream-regulated gene 4 (NDRG4): a candidate tumor suppressor gene and potential biomarker for colorectal cancer. J Natl Cancer Inst 101(13):916–927. https://doi.org/10.1093/jnci/djp131
Xiao Z, Li B, Wang G, Zhu W, Wang Z, Lin J, Xu A, Wang X (2014) Validation of methylation-sensitive high-resolution melting (MS-HRM) for the detection of stool DNA methylation in colorectal neoplasms. Clin Chim Acta 431:154–163. https://doi.org/10.1016/j.cca.2014.01.044
Gyparaki MT, Basdra EK, Papavassiliou AG (2013) DNA methylation biomarkers as diagnostic and prognostic tools in colorectal cancer. J Mol Medi (Berlin, Germany) 91(11):1249–1256. https://doi.org/10.1007/s00109-013-1088-z
Lamb YN, Dhillon S (2017) Epi proColon((R)) 2.0 CE: a blood-based screening test for colorectal cancer. Mol Diagn Ther 21(2):225–232. https://doi.org/10.1007/s40291-017-0259-y
Jin P, Kang Q, Wang X, Yang L, Yu Y, Li N, He YQ, Han X, Hang J, Zhang J, Song L, Han Y, Sheng JQ (2015) Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol 30(5):830–833. https://doi.org/10.1111/jgh.12855
Fu B, Yan P, Zhang S, Lu Y, Pan L, Tang W, Chen S, Chen S, Zhang A, Liu W (2018) Cell-free circulating methylated SEPT9 for noninvasive diagnosis and monitoring of colorectal cancer. Dis Markers 2018:6437104. https://doi.org/10.1155/2018/6437104
Mitchell SM, Ho T, Brown GS, Baker RT, Thomas ML, McEvoy A, Xu ZZ, Ross JP, Lockett TJ, Young GP, LaPointe LC, Pedersen SK, Molloy PL (2016) Evaluation of methylation biomarkers for detection of circulating tumor DNA and application to colorectal cancer. Genes 7(12). https://doi.org/10.3390/genes7120125
Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, Castanos-Velez E, Blumenstein BA, Rosch T, Osborn N, Snover D, Day RW, Ransohoff DF (2014) Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 63(2):317–325. https://doi.org/10.1136/gutjnl-2012-304149
Toth K, Sipos F, Kalmar A, Patai AV, Wichmann B, Stoehr R, Golcher H, Schellerer V, Tulassay Z, Molnar B (2012) Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One 7(9):e46000. https://doi.org/10.1371/journal.pone.0046000
Danese E, Minicozzi AM, Benati M, Montagnana M, Paviati E, Salvagno GL, Lima-Oliveira G, Gusella M, Pasini F, Lippi G, Guidi GC (2015) Comparison of genetic and epigenetic alterations of primary tumors and matched plasma samples in patients with colorectal cancer. PLoS One 10(5):e0126417. https://doi.org/10.1371/journal.pone.0126417
Bartak BK, Kalmar A, Peterfia B, Patai AV, Galamb O, Valcz G, Spisak S, Wichmann B, Nagy ZB, Toth K, Tulassay Z, Igaz P, Molnar B (2017) Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics 12(9):751–763. https://doi.org/10.1080/15592294.2017.1356957
Takeda M, Nagasaka T, Dong-Sheng S, Nishie H, Oka T, Yamada E, Mori Y, Shigeyasu K, Morikawa T, Mizobuchi S, Fujiwara T (2011) Expansion of CpG methylation in the SFRP2 promoter region during colorectal tumorigenesis. Acta Med Okayama 65(3):169–177. https://doi.org/10.18926/AMO/46628
Muller HM, Oberwalder M, Fiegl H, Morandell M, Goebel G, Zitt M, Muhlthaler M, Ofner D, Margreiter R, Widschwendter M (2004) Methylation changes in faecal DNA: a marker for colorectal cancer screening? Lancet 363(9417):1283–1285. https://doi.org/10.1016/S0140-6736(04)16002-9
Huang Z, Li L, Wang J (2007) Hypermethylation of SFRP2 as a potential marker for stool-based detection of colorectal cancer and precancerous lesions. Dig Dis Sci 52(9):2287–2291. https://doi.org/10.1007/s10620-007-9755-y
Huang ZH, Li LH, Yang F, Wang JF (2007) Detection of aberrant methylation in fecal DNA as a molecular screening tool for colorectal cancer and precancerous lesions. World J Gastroenterol 13(6):950–954
Oberwalder M, Zitt M, Wontner C, Fiegl H, Goebel G, Zitt M, Kohle O, Muhlmann G, Ofner D, Margreiter R, Muller HM (2008) SFRP2 methylation in fecal DNA--a marker for colorectal polyps. Int J Color Dis 23(1):15–19. https://doi.org/10.1007/s00384-007-0355-2
Nagasaka T, Tanaka N, Cullings HM, Sun DS, Sasamoto H, Uchida T, Koi M, Nishida N, Naomoto Y, Boland CR, Matsubara N, Goel A (2009) Analysis of fecal DNA methylation to detect gastrointestinal neoplasia. J Natl Cancer Inst 101(18):1244–1258. https://doi.org/10.1093/jnci/djp265
Wang J, Zhang M, Wang P, Zhang C (2016) Detection of CNRIP1, SNCA, SFRP2, and Vimentin methylation in the stool as a biomarker for colorectal cancer. J Gastroenterol Hepatol (Australia) 31(Supplement 3):135. https://doi.org/10.1111/jgh.13540
Lind GE, Raiborg C, Danielsen SA, Rognum TO, Thiis-Evensen E, Hoff G, Nesbakken A, Stenmark H, Lothe RA (2011) SPG20, a novel biomarker for early detection of colorectal cancer, encodes a regulator of cytokinesis. Oncogene 30(37):3967–3978. https://doi.org/10.1038/onc.2011.109
Hibi K, Goto T, Shirahata A, Saito M, Kigawa G, Nemoto H, Sanada Y (2011) Detection of TFPI2 methylation in the serum of colorectal cancer patients. Cancer Lett 311(1):96–100. https://doi.org/10.1016/j.canlet.2011.07.006
Zhang J, Yang S, Xie Y, Chen X, Zhao Y, He D, Li J (2012) Detection of methylated tissue factor pathway inhibitor 2 and human long DNA in fecal samples of patients with colorectal cancer in China. Cancer Epidemiol 36(1):73–77. https://doi.org/10.1016/j.canep.2011.04.006
Zhang H, Zhu YQ, Wu YQ, Zhang P, Qi J (2014) Detection of promoter hypermethylation of Wnt antagonist genes in fecal samples for diagnosis of early colorectal cancer. World J Gastroenterol 20(20):6329–6335. https://doi.org/10.3748/wjg.v20.i20.6329
Ned RM, Melillo S, Marrone M (2011) Fecal DNA testing for colorectal cancer screening: the ColoSure test. PLoS Curr 3:RRN1220. https://doi.org/10.1371/currents.RRN1220
Amiot A, Mansour H, Baumgaertner I, Delchier JC, Tournigand C, Furet JP, Carrau JP, Canoui-Poitrine F, Sobhani I, Marne CRCgoVD (2014) The detection of the methylated Wif-1 gene is more accurate than a fecal occult blood test for colorectal cancer screening. PLoS One 9(7):e99233. https://doi.org/10.1371/journal.pone.0099233
Carmona FJ, Azuara D, Berenguer T, Fernandez A, Biondo S, De Oca J, Rodriguez-Moranta F, Salazar R, Villanueva A, Fraga M, Guardiola J, Capella G, Esteller M, Moreno V (2013) Dna methylation biomarkers for non-invasive diagnostic of colorectal cancer. United European Gastroenterol J 1:A386. https://doi.org/10.1177/2050640613502900
Herbst A, Rahmig K, Stieber P, Philipp A, Jung A, Ofner A, Crispin A, Neumann J, Lamerz R, Kolligs FT (2011) Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol 106(6):1110–1118. https://doi.org/10.1038/ajg.2011.6
Shirahata A, Hibi K (2014) Serum vimentin methylation as a potential marker for colorectal cancer. Anticancer Res 34(8):4121–4125
Itzkowitz SH, Jandorf L, Brand R, Rabeneck L, Schroy PC 3rd, Sontag S, Johnson D, Skoletsky J, Durkee K, Markowitz S, Shuber A (2007) Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol 5(1):111–117. https://doi.org/10.1016/j.cgh.2006.10.006
Baek YH, Chang E, Kim YJ, Kim BK, Sohn JH, Park DI (2009) Stool methylation-specific polymerase chain reaction assay for the detection of colorectal neoplasia in Korean patients. Dis Colon Rectum 52(8):1452–1459; discussion 1459-1463. https://doi.org/10.1007/DCR.0b013e3181a79533
Li M, Chen WD, Papadopoulos N, Goodman SN, Bjerregaard NC, Laurberg S, Levin B, Juhl H, Arber N, Moinova H, Durkee K, Schmidt K, He YP, Diehl F, Velculescu VE, Zhou SB, Diaz LA, Kinzler KW, Markowitz SD, Vogelstein B (2009) Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol 27(9):858–U118. https://doi.org/10.1038/nbt.1559
Itzkowitz S, Brand R, Jandorf L, Durkee K, Millholland J, Rabeneck L, Schroy PC 3rd, Sontag S, Johnson D, Markowitz S, Paszat L, Berger BM (2008) A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol 103(11):2862–2870. https://doi.org/10.1111/j.1572-0241.2008.02088.x
Liu Y, Wu H, Zhou Q, Song Q, Rui J, Zou B, Zhou G (2017) Digital quantification of gene methylation in stool DNA by emulsion-PCR coupled with hydrogel immobilized bead-array. Biosens Bioelectron 92:596–601. https://doi.org/10.1016/j.bios.2016.10.054
Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, Pretlow TP, Lutterbaugh J, Kasturi L, Willson JK, Rao JS, Shuber A, Markowitz SD (2005) Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst 97(15):1124–1132. https://doi.org/10.1093/jnci/dji204
Xue X, Teare MD, Holen I, Zhu YM, Woll PJ (2009) Optimizing the yield and utility of circulating cell-free DNA from plasma and serum. Clin Chim Acta 404(2):100–104. https://doi.org/10.1016/j.cca.2009.02.018
Sozzi G, Roz L, Conte D, Mariani L, Andriani F, Verderio P, Pastorino U (2005) Effects of prolonged storage of whole plasma or isolated plasma DNA on the results of circulating DNA quantification assays. J Natl Cancer Inst 97(24):1848–1850. https://doi.org/10.1093/jnci/dji432
Yuasa Y (2010) Epigenetics in molecular epidemiology of cancer a new scope. Adv Genet 71:211–235. https://doi.org/10.1016/B978-0-12-380864-6.00007-9
Ioannidis JPA, Bossuyt PMM (2017) Waste, leaks, and failures in the biomarker pipeline. Clin Chem 63(5):963–972. https://doi.org/10.1373/clinchem.2016.254649
Lind GE, van Engeland M (2017) Details matter: the role of genomic location and assay standardization in DNA methylation analyses. Epigenomics 9(7):933–935. https://doi.org/10.2217/epi-2017-0053
Code availability
Not applicable.
Funding
Open Access funding provided by NTNU Norwegian University of Science and Technology (incl St. Olavs Hospital - Trondheim University Hospital).
Author information
Authors and Affiliations
Contributions
E.A.L., S.S.B., and F.S. collected, analyzed, and interpreted the data together. E.A.L. wrote the first draft of the manuscript. All authors revised the manuscript and approved the final version of it.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors gave their consent to publication.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Laugsand, E.A., Brenne, S.S. & Skorpen, F. DNA methylation markers detected in blood, stool, urine, and tissue in colorectal cancer: a systematic review of paired samples. Int J Colorectal Dis 36, 239–251 (2021). https://doi.org/10.1007/s00384-020-03757-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00384-020-03757-x