Abstract
Purpose
The purpose of our study is to identify potential biomarkers of hepatoblastoma (HB) and further explore the pathogenesis of it.
Methods
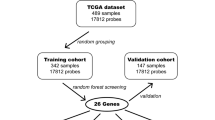
Differentially expressed genes (DEGs) were incorporated into the combined random forest and artificial neural network diagnosis model to screen candidate genes for HB. Gene set enrichment analysis (GSEA) was used to analyze the ARHGEF2. Student’s t test was performed to evaluate the difference of tumor-infiltrating immune cells (TIICs) between normal and HB samples. Spearson correlation analysis was used to calculate the correlation between ARHGEF2 and TIICs.
Results
ARHGEF2, TCF3, TMED3, STMN1 and RAVER2 were screened by the new model. The GSEA of ARHGEF2 included cell cycle pathway and antigen processing presenting pathway. There were significant differences in the composition of partial TIICs between HB and normal samples (p < 0.05). ARHGEF2 was significantly correlated with memory B cells (Cor = 0.509, p < 0.05).
Conclusion
These 5 candidate genes contribute to the molecular diagnosis and targeted therapy of HB. And we found “ARHGEF2–RhoA–Cyclin D1/CDK4/CDK6–EF2” is a key mechanism regulating cell cycle pathway in HB. This will be helpful in the treatment of HB. The occurrence of HB is related to abnormal TIICs. We speculated that memory B cells play an important role in HB.






Similar content being viewed by others
References
Liu L, Wang J, Sun G et al (2019) m6A mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol Cancer 18:188. https://doi.org/10.1186/s12943-019-1119-7
Hager J, Sergi CM (2021) Hepatoblastoma. In: Sergi CM (ed) Liver cancer [Internet]. Exon publications, Brisbane. Chapter 8. https://doi.org/10.36255/exonpublications.livercancer.2021.ch8
Hafberg E, Borinstein SC, Alexopoulos SP (2019) Contemporary management of hepatoblastoma. Curr Opin Organ Transplant 24(2):113–117. https://doi.org/10.1097/MOT.0000000000000618
Edgar R, Lash A (2002). The Gene Expression Omnibus (GEO): a gene expression and hybridization repository. http://www.icgeb.res.in/whotdr/cd1/PreCourseReading/NCBI_Handbook2/ch6d1.pdf
Valanejad L, Cast A, Wright M et al (2018) PARP1 activation increases expression of modified tumor suppressors and pathways underlying development of aggressive hepatoblastoma. Commun Biol 1:67. https://doi.org/10.1038/s42003-018-0077-8
Hiyama E, Ueda Y, Kurihara S et al (2021) Gene expression profiling in hepatoblastoma cases of the Japanese Study Group for Pediatric Liver Tumors-2 (JPLT-2) trial. European J Mol Cancer. https://doi.org/10.31487/j.EJMC.2018.01.003
Carrillo-Reixach J, Torrens L, Simon-Coma M et al (2020) Epigenetic footprint enables molecular risk stratification of hepatoblastoma with clinical implications. J Hepatol 73(2):328–341. https://doi.org/10.1016/j.jhep.2020.03.025
Ritchie ME, Phipson B, Wu D et al (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. https://doi.org/10.1093/nar/gkv007
Lê S, Josse J, Husson F (2008) FactoMineR : an R package for multivariate analysis. J Stat Soft. https://doi.org/10.18637/jss.v025.i01
Wickham H. (2016) ggplot2: Elegant graphics for data analysis. Springer-verlag New York. https://ggplot2.tidyverse.orghttps://ggplot2.tidyverse.org
Kolde R. (2019) pheatmap: Pretty Heatmaps. https://CRAN.R-project.org/package=pheatmap. Accessed 6 May 2022
Wu T, Hu E, Xu S et al (2021) clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2(3):100141. https://doi.org/10.1016/j.xinn.2021.100141
Doms A, Schroeder M (2005) GoPubMed: exploring PubMed with the gene ontology. Nucleic Acids Res 33:W783–W786. https://doi.org/10.1093/nar/gki470
Kanehisa M, Goto S, Kawashima S, Nakaya A (2002) The KEGG databases at genomenet. Nucleic Acids Res 30(1):42–46. https://doi.org/10.1093/nar/30.1.42
Liaw A, Wiener M (2002). Classification and Regression by randomForest. R News 2(3), 18-22. https://CRAN.R-project.org/doc/Rnews/
Robin X, Turck N, Hainard A et al (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77. https://doi.org/10.1186/1471-2105-12-77
Günther F, Fritsch S (2010) Neuralnet: training of neural networks. The R Journal 2(1):30. https://doi.org/10.32614/RJ-2010-006
Kassambara A. (2020) ggpubr: “ggplot2” Based Publication Ready Plots. Published online June 27, 2020. https://CRAN.R-project.org/package=ggpubr. Accessed 14 May 2022
Luo W, Brouwer C (2013) Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29(14):1830–1831. https://doi.org/10.1093/bioinformatics/btt285
Hänzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 14(1):7. https://doi.org/10.1186/1471-2105-14-7
Czauderna P, Garnier H (2018) Hepatoblastoma: current understanding, recent advances, and controversies. F1000Res 7:53. https://doi.org/10.12688/f1000research.12239.1
Sha YL, Liu S, Yan WW, Dong B (2019) Wnt/β-catenin signaling as a useful therapeutic target in hepatoblastoma. Biosci Rep 39(9):BSR20192466. https://doi.org/10.1042/BSR20192466
Bao P, Yokobori T, Altan B et al (2017) High STMN1 expression is associated with cancer progression and chemo-resistance in lung squamous cell carcinoma. Ann Surg Oncol 24(13):4017–4024. https://doi.org/10.1245/s10434-017-6083-0
Yan L, Dong X, Gao J et al (2018) A novel rapid quantitative method reveals stathmin-1 as a promising marker for esophageal squamous cell carcinoma. Cancer Med 7(5):1802–1813. https://doi.org/10.1002/cam4.1449
Zhou Q, Ching AKK, Leung WKC et al (2011) Novel therapeutic potential in targeting microtubules by nanoparticle albumin-bound paclitaxel in hepatocellular carcinoma. Int J Oncol 38(3):721–731. https://doi.org/10.3892/ijo.2011.902
Nagata T, Takahashi Y, Ishii Y et al (2003) Transcriptional profiling in hepatoblastomas using high-density oligonucleotide DNA array. Cancer Genet Cytogenet 145(2):152–160. https://doi.org/10.1016/s0165-4608(03)00065-7
Pu XY, Zheng DF, Lv T, Zhou YJ, Yang JY, Jiang L (2022) Overexpression of transcription factor 3 drives hepatocarcinoma development by enhancing cell proliferation via activating Wnt signaling pathway. Hepatobiliary Pancreat Dis Int. https://doi.org/10.1016/j.hbpd.2022.01.003
Zheng H, Yang Y, Han J et al (2016) TMED3 promotes hepatocellular carcinoma progression via IL-11/STAT3 signaling. Sci Rep 6:37070. https://doi.org/10.1038/srep37070
Bartoletti-Stella A, Gasparini L, Giacomini C et al (2015) Messenger RNA processing is altered in autosomal dominant leukodystrophy. Hum Mol Genet 24(10):2746–2756. https://doi.org/10.1093/hmg/ddv034
Bouzid D, Fourati H, Amouri A et al (2012) Association of the RAVER2 gene with increased susceptibility for ulcerative colitis. Hum Immunol 73(7):732–735. https://doi.org/10.1016/j.humimm.2012.04.018
Birukova AA, Adyshev D, Gorshkov B, Bokoch GM, Birukov KG, Verin AD (2006) GEF-H1 is involved in agonist-induced human pulmonary endothelial barrier dysfunction. Am J Physiol Lung Cell Mol Physiol 290(3):L540-548. https://doi.org/10.1152/ajplung.00259.2005
Cullis J, Meiri D, Sandi MJ et al (2014) The RhoGEF GEF-H1 is required for oncogenic RAS signaling via KSR-1. Cancer Cell 25(2):181–195. https://doi.org/10.1016/j.ccr.2014.01.025
Nalbant P, Chang YC, Birkenfeld J, Chang ZF, Bokoch GM (2009) Guanine nucleotide exchange factor-H1 regulates cell migration via localized activation of RhoA at the leading edge. Mol Biol Cell 20(18):4070–4082. https://doi.org/10.1091/mbc.E09-01-0041
Cao J, Yang T, Tang D, Zhou F, Qian Y, Zou X (2019) Increased expression of GEF-H1 promotes colon cancer progression by RhoA signaling. Pathol Res Pract 215(5):1012–1019. https://doi.org/10.1016/j.prp.2019.02.008
Wang S, Gao S, Zeng Y et al (2022) N6-methyladenosine reader YTHDF1 promotes ARHGEF2 translation and RhoA signaling in colorectal cancer. Gastroenterology 162(4):1183–1196. https://doi.org/10.1053/j.gastro.2021.12.269
Joo E, Olson MF (2021) Regulation and functions of the RhoA regulatory guanine nucleotide exchange factor GEF-H1. Small GTPases 12(5–6):358–371. https://doi.org/10.1080/21541248.2020.1840889
Cheng IK, Tsang BC, Lai KP et al (2012) GEF-H1 over-expression in hepatocellular carcinoma promotes cell motility via activation of RhoA signalling. J Pathol 228(4):575–585. https://doi.org/10.1002/path.4084
Chen H, Gao F, He M et al (2019) Long-read RNA sequencing identifies alternative splice variants in hepatocellular carcinoma and tumor-specific isoforms. Hepatology 70(3):1011–1025. https://doi.org/10.1002/hep.30500
Nakao Y, Nakagawa S, Yamashita ichi Y et al (2021) High ARHGEF2 (GEF-H1) expression is associated with poor prognosis via cell cycle regulation in patients with pancreatic cancer. Ann Surg Oncol 28(8):4733–4743. https://doi.org/10.1245/s10434-020-09383-9
Nie M, Aijaz S, Leefa Chong San IV, Balda MS, Matter K (2009) The Y-box factor ZONAB/DbpA associates with GEF-H1/Lfc and mediates Rho-stimulated transcription. EMBO Rep 10(10):1125–1131. https://doi.org/10.1038/embor.2009.182
Montalto FI, De Amicis F (2020) Cyclin D1 in cancer: a molecular connection for cell cycle control, adhesion and invasion in tumor and stroma. Cells 9(12):2648. https://doi.org/10.3390/cells9122648
Zhang Y, Zhang T, Yin Q, Luo H (2021) Development and validation of genomic and epigenomic signatures associated with tumor immune microenvironment in hepatoblastoma. BMC Cancer 21:1156. https://doi.org/10.1186/s12885-021-08893-3
Calvisi DF, Solinas A (2020) Hepatoblastoma: current knowledge and promises from preclinical studies. Transl Gastroenterol Hepatol 5:42. https://doi.org/10.21037/tgh.2019.12.03
Jin J, Jin J, Woodfield SE et al (2019) Targeting LRH-1 in hepatoblastoma cell lines causes decreased proliferation. Oncol Rep 41(1):143–153. https://doi.org/10.3892/or.2018.6793
Acknowledgements
We acknowledge the Tianjin Science and Technology Program and the Natural Science Foundation of Xinjiang Uygur Autonomous Region for research support.
Author information
Authors and Affiliations
Contributions
SWL and TFL: conceived and designed the study. SWL and RFZ: collecting and analyzing data, drafting the manuscript. QPZ and JHZ: read and approved the final manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethical approval
Not applicable.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
383_2022_5255_MOESM1_ESM.tif
Supplementary file1 Standardization for the datasets. A The boxplot before standardization of the new dataset. B The boxplot after standardization of the new data set. C The boxplot before standardization of the GSE133039 dataset. D The boxplot after standardization of the GSE133039 dataset. The X-axis represents the sample. The Y-axis represents the value of gene expression. The black horizontal line in each box represents the median of the data, and the median represents the standardized gene expression value (TIF 51212 KB)
383_2022_5255_MOESM2_ESM.tif
Supplementary file 2 PCA for the datasets. A PCA for the classifications of normal samples and hepatoblastoma samples in the new dataset. B PCA for the classifications of normal samples and hepatoblastoma samples in the GSE133039 dataset (TIF 128615 KB)
383_2022_5255_MOESM6_ESM.tif
Supplementary file 6 Total of 321 DEGs identified from 2 datasets (6623 in the GSE133039 dataset and 554 in the new dataset) (TIF 14242 KB)
383_2022_5255_MOESM7_ESM.tif
Supplementary file 7 Volcano maps and heat maps of DEGs in two datasets. Up-regulated DEGs is red, reduced DEGs are shown in blue, non-DEGs are indicated in gray. A Volcano maps for the new dataset. B Volcano maps for GSE133039 dataset. C Heat maps for the new dataset. D Heat maps for GSE133039 dataset (TIF 59161 KB)
383_2022_5255_MOESM10_ESM.tif
Supplementary file 10 Functional enrichment analysis for DEGs. A GO enrichment bubble diagram of DEGs (Top 5). B KEGG pathway enrichment bubble map of DEGs (Top 15). C Circular graph of KEGG pathway of DEGs (Top 15). D Interaction network of 15 KEGG pathways (TIF 49787 KB)
383_2022_5255_MOESM14_ESM.tif
Supplementary file 14 Construction of ceRNA network. mRNAs, miRNAs and lncRNAs were represented by purple circle node, yellow V-shaped node and green diamond node, respectively (TIF 44049 KB)
383_2022_5255_MOESM16_ESM.tif
Supplementary file 16 Unsupervised clustering of 5 candidate genes. A Clustering of the 5 candidate genes in new dataset. B Clustering of the 5 candidate genes in GSE133039 dataset (TIF 57900 KB)
383_2022_5255_MOESM17_ESM.tif
Supplementary file 17 The expression differences of the 5 candidate genes between tumor and normal samples. The expression of A ARHGEF2, B TCF3, C TMED3, D STMN1, E RAVER2 was significantly up-regulated in the tumor samples. *** P<0.001, ** P<0.01, * P<0.05 (TIF 35085 KB)
383_2022_5255_MOESM21_ESM.tif
Supplementary file 21 Correlation between ARHGEF2 and immune cells. A-O Correlation between ARHGEF2 and activated CD4 T cell, activated CD8 T cell, activated dendritic cell, CD56bright natural killer cel, CD56dim natural killer cell, central memory CD4 T cell, eosinophil, gamma delta T cell, memory B cell, monocyte, neutrophil, plasmacytoid dendritic cell, regulatory T cell , T follicular helper cell and type 2 T helper cell (TIF 217306 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, S., Zheng, Q., Zhang, R. et al. Construction of a combined random forest and artificial neural network diagnosis model to screening potential biomarker for hepatoblastoma. Pediatr Surg Int 38, 2023–2034 (2022). https://doi.org/10.1007/s00383-022-05255-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00383-022-05255-3