Abstract
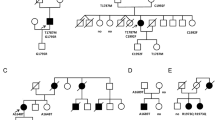
We found that a female infant presenting with left bundle branch block and left ventricular noncompaction carries uninvestigated gene mutations HCN4(G811E), SCN5A(L1988R), DMD(S2384Y), and EMD(R203H). Here, we explored the possible pathogenicity of HCN4(G811E), which results in a G811E substitution in hyperpolarization-activated cyclic nucleotide-gated channel 4, the main subunit of the cardiac pacemaker channel. Voltage-clamp measurements in a heterologous expression system of HEK293T cells showed that HCN4(G811E) slightly reduced whole-cell HCN4 channel conductance, whereas it did not affect the gating kinetics, unitary conductance, or cAMP-dependent modulation of voltage-dependence. Immunocytochemistry and immunoblot analysis showed that the G811E mutation did not impair the membrane trafficking of the channel subunit in the heterologous expression system. These findings indicate that HCN4(G811E) may not be a monogenic factor to cause the cardiac disorders.






Similar content being viewed by others
References
Shi W, Wymore R, Yu H, Wu J, Wymore RT, Pan Z, Robinson RB, Dixon JE, McKinnon D, Cohen IS (1999) Distribution and prevalence of hyperpolarization-activated cation channel (HCN) mRNA expression in cardiac tissues. Circ Res 85:e1–e6
Verkerk AO, Wilders R (2014) Pacemaker activity of the human sinoatrial node: effects of HCN4 mutations on the hyperpolarization-activated current. Europace 16:384–395
DiFrancesco D (2015) HCN4, sinus bradycardia and atrial fibrillation. Arrhythm Electrophysiol Rev 4:9–13
Duhme N, Schweizer PA, Thomas D, Becker R, Schroter J, Barends TR, Schlichting I, Draguhn A, Bruehl C, Katus HA, Koenen M (2013) Altered HCN4 channel C-linker interaction is associated with familial tachycardia-bradycardia syndrome and atrial fibrillation. Eur Heart J 34:2768–2775
Schweizer PA, Duhme N, Thomas D, Becker R, Zehelein J, Draguhn A, Bruehl C, Katus HA, Koenen M (2010) cAMP sensitivity of HCN pacemaker channels determines basal heart rate but is not critical for autonomic rate control. Circ Arrhythm Electrophysiol 3:542–552
Schulze-Bahr E, Neu A, Friederich P, Kaupp UB, Breithardt G, Pongs O, Isbrandt D (2003) Pacemaker channel dysfunction in a patient with sinus node disease. J Clin Investig 111:1537–1545
Hategan L, Csanyi B, Ordog B, Kakonyi K, Tringer A, Kiss O, Orosz A, Saghy L, Nagy I, Hegedus Z, Rudas L, Szell M, Varro A, Forster T, Sepp R (2017) A novel ‘splice site’ HCN4 Gene mutation, c.1737 + 1 G>T, causes familial bradycardia, reduced heart rate response, impaired chronotropic competence and increased short-term heart rate variability. Int J Cardiol 241:364–372
Milano A, Vermeer AM, Lodder EM, Barc J, Verkerk AO, Postma AV, van der Bilt IA, Baars MJ, van Haelst PL, Caliskan K, Hoedemaekers YM, Le Scouarnec S, Redon R, Pinto YM, Christiaans I, Wilde AA, Bezzina CR (2014) HCN4 mutations in multiple families with bradycardia and left ventricular noncompaction cardiomyopathy. J Am Coll Cardiol 64:745–756
Schweizer PA, Schröter J, Greiner S, Haas J, Yampolsky P, Mereles D, Buss SJ, Seyler C, Bruehl C, Draguhn A, Koenen M, Meder B, Katus HA, Thomas D (2014) The symptom complex of familial sinus node dysfunction and myocardial noncompaction is associated with mutations in the HCN4 channel. J Am Coll Cardiol 64:757–767
Liang X, Evans SM, Sun Y (2015) Insights into cardiac conduction system formation provided by HCN4 expression. Trends Cardiovasc Med 25:1–9
Ichida F, Tsubata S, Bowles KR, Haneda N, Uese K, Miyawaki T, Dreyer WJ, Messina J, Li H, Bowles NE, Towbin JA (2001) Novel gene mutations in patients with left ventricular noncompaction or Barth syndrome. Circulation 103:1256–1263
Hata Y, Kinoshita K, Mizumaki K, Yamaguchi Y, Hirono K, Ichida F, Takasaki A, Mori H, Nishida N (2016) Postmortem genetic analysis of sudden unexplained death syndrome under 50 years of age: a next-generation sequencing study. Heart Rhythm 13:1544–1551
Yamaguchi Y, Nishide K, Kato M, Hata Y, Mizumaki K, Kinoshita K, Nonobe Y, Tabata T, Sakamoto T, Kataoka N, Nakatani Y, Ichida F, Mori H, Fukurotani K, Inoue H, Nishida N (2014) Glycine/serine polymorphism at position 38 influences KCNE1 subunit’s modulatory actions on rapid and slow delayed rectifier K+ currents. Circ J 78:610–618
Maekawa K, Saito Y, Ozawa S, Adachi-Akahane S, Kawamoto M, Komamura K, Shimizu W, Ueno K, Kamakura S, Kamatani N, Kitakaze M, Sawada J (2005) Genetic polymorphisms and haplotypes of the human cardiac sodium channel alpha subunit gene (SCN5A) in Japanese and their association with arrhythmia. Ann Hum Genet 69:413–428
Lee YS, Olaopa MA, Jung BC, Lee SH, Shin DG, Park HS, Cho Y, Han SM, Lee MH, Kim YN (2016) Genetic variation of SCN5A in Korean patients with sick sinus syndrome. Korean Circ J 46:63–71
Hussein A, Karimianpour A, Collier P, Krasuski RA (2015) Isolated noncompaction of the left ventricle in adults. J Am Coll Cardiol 66:578–585
Ohno S, Omura M, Kawamura M, Kimura H, Itoh H, Makiyama T, Ushinohama H, Makita N, Horie M (2014) Exon 3 deletion of RYR2 encoding cardiac ryanodine receptor is associated with left ventricular non-compaction. Europace 16:1646–1654
Towbin JA, Lorts A, Jefferies JL (2015) Left ventricular non-compaction cardiomyopathy. Lancet 386(9995):813–825
Lolicato M, Bucchi A, Arrigoni C, Zucca S, Nardini M, Schroeder I, Simmons K, Aquila M, DiFrancesco D, Bolognesi M, Schwede F, Kashin D, Fishwick CW, Johnson AP, Thiel G, Moroni A (2014) Cyclic dinucleotides bind the C-linker of HCN4 to control channel cAMP responsiveness. Nat Chem Biol 10:457–462
DiFrancesco D (2010) The role of the funny current in pacemaker activity. Circ Res 106:434–446
Liao Z, Lockhead D, St Clair JR, Larson ED, Wilson CE, Proenza C (2012) Cellular context and multiple channel domains determine cAMP sensitivity of HCN4 channels: ligand-independent relief of autoinhibition in HCN4. J Gen Physiol 140:557–566
DiFrancesco D, Tortora P (1991) Direct activation of cardiac pacemaker channels by intracellular cyclic AMP. Nature 351:145–147
DiFrancesco D (1999) Dual allosteric modulation of pacemaker (f) channels by cAMP and voltage in rabbit SA node. J Physiol 515:367–376
Laish-Farkash A, Glikson M, Brass D, Marek-Yagel D, Pras E, Dascal N, Antzelevitch C, Nof E, Reznik H, Eldar M, Luria D (2010) A novel mutation in the HCN4 gene causes symptomatic sinus bradycardia in Moroccan Jews. J Cardiovasc Electrophysiol 21:1365–1372
Macri V, Mahida SN, Zhang ML, Sinner MF, Dolmatova EV, Tucker NR, McLellan M, Shea MA, Milan DJ, Lunetta KL, Benjamin EJ, Ellinor PT (2014) A novel trafficking-defective HCN4 mutation is associated with early-onset atrial fibrillation. Heart Rhythm 11:1055–1062
Nof E, Luria D, Brass D, Marek D, Lahat H, Reznik-Wolf H, Pras E, Dascal N, Eldar M, Glikson M (2007) Point mutation in the HCN4 cardiac ion channel pore affecting synthesis, trafficking, and functional expression is associated with familial asymptomatic sinus bradycardia. Circulation 116:463–470
Baruscotti M, Barbuti A, Bucchi A (2010) The cardiac pacemaker current. J Mol Cell Cardiol 48:55–64
Ferlini A, Neri M, Gualandi F (2013) The medical genetics of dystrophinopathies: molecular genetic diagnosis and its impact on clinical practice. Neuromuscul Disord 23:4–14
Arbustini E, Weidemann F, Hall JL (2014) Left ventricular noncompaction: a distinct cardiomyopathy or a trait shared by different cardiac diseases? J Am Coll Cardiol 64:1840–1850
Kimura K, Morita H, Daimon M, Kawata T, Nakao T, Lee SL, Hirokawa M, Ebihara A, Nakajima T, Ozawa T, Yonemochi Y, Aida I, Motoyoshi Y, Mikata T, Uchida I, Komori T, Kitao R, Nagata T, Takeda S, Komaki H, Segawa K, Takenaka K, Komuro I (2015) Prognostic impact of venous thromboembolism in patients with Duchenne muscular dystrophy: prospective multicenter 5-year cohort study. Int J Cardiol 191:178–180
Parent JJ, Moore RA, Taylor MD, Towbin JA, Jefferies JL (2015) Left ventricular noncompaction cardiomyopathy in Duchenne muscular dystrophy carriers. J Cardiol Cases 11:7–9
Meinke P, Nguyen TD, Wehnert MS (2011) The LINC complex and human disease. Biochem Soc Trans 39:1693–1697
Yuan J, Xue B (2015) Role of structural flexibility in the evolution of emerin. J Theor Biol 385:102–111
Parmar MS, Parmar KS (2012) Emery-Dreifuss humeroperoneal muscular dystrophy: cardiac manifestations. Can J Cardiol 28(4):516.e1–516.e3
Monserrat L, Hermida-Prieto M, Fernandez X, Rodríguez I, Dumont C, Cazón L, Cuesta MG, Gonzalez-Juanatey C, Peteiro J, Álvarez N, Penas-Lado M, Castro-Beiras A (2007) Mutation in the alpha-cardiac actin gene associated with apical hypertrophic cardiomyopathy, left ventricular non-compaction, and septal defects. Eur Heart J 28:1953–1961
Girolami F, Iascone M, Tomberli B, Bardi S, Benelli M, Marseglia G, Pescucci C, Pezzoli L, Sana ME, Basso C, Marziliano N, Merlini PA, Fornaro A, Cecchi F, Torricelli F, Olivotto I (2014) Novel alpha-actinin 2 variant associated with familial hypertrophic cardiomyopathy and juvenile atrial arrhythmias: a massively parallel sequencing study. Circ Cardiovasc Genet 7:741–750
Stöllberger C, Finsterer J, Blazek G, Bittner RE (1996) Left ventricular non-compaction in a patient with Becker’s muscular dystrophy. Heart 76:380
Williams T, Machann W, Kuhler L, Hamm H, Muller-Hocker J, Zimmer M, Ertl G, Ritter O, Beer M, Schonberger J (2011) Novel desmoplakin mutation: juvenile biventricular cardiomyopathy with left ventricular non-compaction and acantholytic palmoplantar keratoderma. Clin Res Cardiol 100:1087–1093
Blinder JJ, Martinez HR, Craigen WJ, Belmont J, Pignatelli RH, Jefferies JL (2011) Noncompaction of the left ventricular myocardium in a boy with a novel chromosome 8p23.1 deletion. Am J Med Genet A 155A:2215–2220
Vatta M, Mohapatra B, Jimenez S, Sanchez X, Faulkner G, Perles Z, Sinagra G, Lin JH, Vu TM, Zhou Q, Bowles KR, Di Lenarda A, Schimmenti L, Fox M, Chrisco MA, Murphy RT, McKenna W, Elliott P, Bowles NE, Chen J, Valle G, Towbin JA (2003) Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J Am Coll Cardiol 42:2014–2027
Liu Z, Shan H, Huang J, Li N, Hou C, Pu J (2016) A novel lamin A/C gene missense mutation (445 V > E) in immunoglobulin-like fold associated with left ventricular non-compaction. Europace 18:617–822
Wessels MW, Herkert JC, Frohn-Mulder IM, Dalinghaus M, van den Wijngaard A, de Krijger RR, Michels M, de Coo IF, Hoedemaekers YM, Dooijes D (2015) Compound heterozygous or homozygous truncating MYBPC3 mutations cause lethal cardiomyopathy with features of noncompaction and septal defects. Eur J Hum Genet 23:922–928
Hoedemaekers YM, Caliskan K, Majoor-Krakauer D, van de Laar I, Michels M, Witsenburg M, ten Cate FJ, Simoons ML, Dooijes D (2007) Cardiac beta-myosin heavy chain defects in two families with non-compaction cardiomyopathy: linking non-compaction to hypertrophic, restrictive, and dilated cardiomyopathies. Eur Heart J 28:2732–2737
Ouyang P, Saarel E, Bai Y, Luo C, Lv Q, Xu Y, Wang F, Fan C, Younoszai A, Chen Q, Tu X, Wang QK (2011) A de novo mutation in NKX2.5 associated with atrial septal defects, ventricular noncompaction, syncope and sudden death. Clin Chim Acta 412:170–175
Hoedemaekers YM, Caliskan K, Michels M, Frohn-Mulder I, van der Smagt JJ, Phefferkorn JE, Wessels MW, ten Cate FJ, Sijbrands EJ, Dooijes D, Majoor-Krakauer DF (2010) The importance of genetic counseling, DNA diagnostics, and cardiologic family screening in left ventricular noncompaction cardiomyopathy. Circ Cardiovasc Genet 3:232–239
Shan L, Makita N, Xing Y, Watanabe S, Futatani T, Ye F, Saito K, Ibuki K, Watanabe K, Hirono K, Uese K, Ichida F, Miyawaki T, Origasa H, Bowles NE, Towbin JA (2008) SCN5A variants in Japanese patients with left ventricular noncompaction and arrhythmia. Mol Genet Metab 93:468–474
Ronvelia D, Greenwood J, Platt J, Hakim S, Zaragoza MV (2012) Intrafamilial variability for novel TAZ gene mutation: Barth syndrome with dilated cardiomyopathy and heart failure in an infant and left ventricular noncompaction in his great-uncle. Mol Genet Metab 107:428–432
Kapadia R, Choudhary P, Collins N, Celermajer D, Puranik R (2016) Left ventricular non-compaction in Holt–Oram syndrome. Heart Lung Circ 25:626–630
Luedde M, Ehlermann P, Weichenhan D, Will R, Zeller R, Rupp S, Müller A, Steen H, Ivandic BT, Ulmer HE, Kern M, Katus HA, Frey N (2010) Severe familial left ventricular non-compaction cardiomyopathy due to a novel troponin T (TNNT2) mutation. Cardiovasc Res 86:452–460
Chang B, Nishizawa T, Furutani M, Fujiki A, Tani M, Kawaguchi M, Ibuki K, Hirono K, Taneichi H, Uese K, Onuma Y, Bowles NE, Ichida F, Inoue H, Matsuoka R, Miyawaki T, Noncompaction Study C (2011) Identification of a novel TPM1 mutation in a family with left ventricular noncompaction and sudden death. Mol Genet Metab 102:200–206
Graham TP Jr, Jarmakani JM, Canent RV Jr, Morrow MN (1971) Left heart volume estimation in infancy and childhood. Reevaluation of methodology and normal values. Circulation 43:895–904
Acknowledgements
We thank the gene sample provider and her family for their kind cooperation and Kohki Nishide, M.Eng., Hiroyuki Takahashi, M.Eng., and Nozomi Hisajima, M.Eng. for their technical assistance. This study was partly supported by JSPS KAKENHI (Grant numbers JP24590852, JP15K08867 to YH; JP26430012 to TT), AMED (Grant number 15dk020710h0002) to TT, and Presidential Discretionary Funds, University of Toyama 2014 to NN.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None for any authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
380_2018_1116_MOESM1_ESM.eps
Supplementary material 1 Supplemental Fig. 1 Flow of the in silico analysis. The variations identified in NGS were screened by the three-step procedure depicted in the flow chart (EPS 2989 kb)
380_2018_1116_MOESM2_ESM.eps
Supplementary material 2 Supplemental Fig. 2 Direct sequencing confirmed the mutations identified using NGS. Partial DNA sequences of the labeled genes from the proband’s family. The corresponding amino acids are shown above each codon. Integer denotes the position of the first nucleotide or amino acid of each partial sequence (EPS 5142 kb)
380_2018_1116_MOESM3_ESM.eps
Supplementary material 3 Supplemental Fig. 3 The L1988R mutation did not change the density or kinetics of SCN5A channel current. a Representative current responses of cells expressing wild-type SCN5A channel or SCN5A(L1988R) channel (WT and LR, respectively) to the voltage stimuli schematically shown above. Note a similarity in the rising and decaying time-courses between the WT and mutant channel currents. Whole-cell recordings were made as described in Kinoshita et al. (Heart Rhythm 13:1113–20, 2016). b Mean peak current density–voltage plots of the channel currents. n, 20 cells for SCN5A(WT) and 23 cells for SCN5A(L1988R). c Mean whole-cell conductance mediated by the channels estimated from the slope of the line fitted to the current density–voltage relation for each cell. d Relative g–V plots of the channel currents. The data were taken from the same cells as in b (EPS 2594 kb)
Rights and permissions
About this article
Cite this article
Yokoyama, R., Kinoshita, K., Hata, Y. et al. A mutant HCN4 channel in a family with bradycardia, left bundle branch block, and left ventricular noncompaction. Heart Vessels 33, 802–819 (2018). https://doi.org/10.1007/s00380-018-1116-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00380-018-1116-6