Abstract
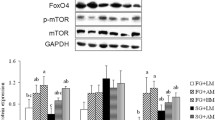
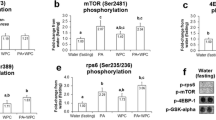
During periods of negative energy balance, mobilization of muscle is a physiologic process providing energy and amino acids. This is important in transition dairy cows experiencing negative energy and protein balance postpartum. Overconsumption of energy during late pregnancy affects resting glucose and insulin concentrations peripartum and increases the risk for hyperketonemia postpartum, but the effects on muscle tissue are not fully understood. Skeletal muscle accounts for the majority of insulin-dependent glucose utilization in ruminants. Our objective was to study peripartal skeletal muscle insulin signaling as well as muscle accretion and atrophy in cows with excess energy consumption prepartum. Skeletal muscle biopsies were obtained 28 and 10 days prepartum, as well as 4 and 21 days postpartum from 24 Holstein cows. Biopsies were taken immediately before and 60 min after intravenous glucose challenge causing endogenous release of insulin. Gene expression of IGF-1, myostatin, and atrogin-1, as well as immunoblot analysis of atrogin-1, muRF1, ubiquitinated proteins, LC3, and phosphorylation of AKT, ERK and mTORC1 substrate 4EBP1 was performed. Excess energy consumption in late pregnancy did not lead to changes in insulin-dependent molecular regulation of muscle accretion or atrophy compared with the controlled energy group. In both groups, phosphorylation of AKT and mTORC1 substrate was significantly decreased postpartum whereas proteasome activity and macroautopagy were upregulated. This study showed that in addition to the proteasome pathway of muscle atrophy, macroautophagy is upregulated in postpartum negative energy and protein balance regardless of dietary energy strategy prepartum and was higher in cows overfed energy throughout the study period.







Similar content being viewed by others
Abbreviations
- AKT:
-
Protein kinase B
- AUC:
-
Area under the curve
- BHB:
-
β-Hydroxy-butyrate
- DMI:
-
Dry matter intake
- 4EBP1:
-
Translation repressor protein 4E-BP1
- EIF3K:
-
Eukaryotic translation initiation factor 3, subunit K
- ERK:
-
Mitogen activated protein kinase/extracellular signal-regulated kinase
- IGF-1:
-
Insulin-like growth factor 1
- IVGTT:
-
Intravenous glucose tolerance test
- LC3:
-
Microtubule-associated protein 1 light chain 3
- MP:
-
Metabolizable protein
- mTORC1:
-
Mammalian target of rapamycin complex 1
- muRF1:
-
Muscle RING-finger protein-1
- PVDF:
-
Polyvinylidene difluoride
- TMR:
-
Total mixed ration
References
Appuhamy JA, Bell AL, Nayananjalie WA, Escobar J, Hanigan MD (2011) Essential amino acids regulate both initiation and elongation of mRNA translation independent of insulin in MAC-T cells and bovine mammary tissue slices. J Nutr 141(6):1209–1215. doi:10.3945/jn.110.136143
Bauman DE, Currie WB (1980) Partitioning of nutrients during pregnancy and lactation: a review of mechanisms involving homeostasis and homeorhesis. J Dairy Sci 63(9):1514–1529
Bell AW (1995) Regulation of organic nutrient metabolism during transition from late pregnancy to early lactation. J Anim Sci 73(9):2804–2819
Bell AW, Burhans WS, Overton TR (2000) Protein nutrition in late pregnancy, maternal protein reserves and lactation performance in dairy cows. Proc Nutr Soc 59(1):119–126
Bonaldo P, Sandri M (2013) Cellular and molecular mechanisms of muscle atrophy. Dis Model Mech 6(1):25–39. doi:10.1242/dmm.010389
Bonnet M, Bernard L, Bes S, Leroux C (2013) Selection of reference genes for quantitative real-time PCR normalisation in adipose tissue, muscle, liver and mammary gland from ruminants. Animal 7(8):1344–1353. doi:10.1017/S1751731113000475
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Carbone JW, McClung JP, Pasiakos SM (2012) Skeletal muscle responses to negative energy balance: effects of dietary protein. Adv Nutr 3(2):119–126. doi:10.3945/an.111.001792
Cardoso FC, Sears W, LeBlanc SJ, Drackley JK (2011) Technical note: comparison of 3 methods for analyzing areas under the curve for glucose and nonesterified fatty acids concentrations following epinephrine challenge in dairy cows. J Dairy Sci 94(12):6111–6115. doi:10.3168/jds.2011-4627
Cecconi F, Levine B (2008) The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 15(3):344–357. doi:10.1016/j.devcel.2008.08.012
Chibisa GE, Gozho GN, Van Kessel AG, Olkowski AA, Mutsvangwa T (2008) Effects of peripartum propylene glycol supplementation on nitrogen metabolism, body composition, and gene expression for the major protein degradation pathways in skeletal muscle in dairy cows. J Dairy Sci 91(9):3512–3527. doi:10.3168/jds.2007-0920
Doepel L, Lapierre H, Kennelly JJ (2002) Peripartum performance and metabolism of dairy cows in response to prepartum energy and protein intake. J Dairy Sci 85(9):2315–2334. doi:10.3168/jds.S0022-0302(02)74312-9
Glass DJ (2003) Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat Cell Biol 5(2):87–90. doi:10.1038/ncb0203-87
Glass DJ (2010) PI3 kinase regulation of skeletal muscle hypertrophy and atrophy. Curr Top Microbiol Immunol 346:267–278. doi:10.1007/82_2010_78
Huang K, Fingar DC (2014) Growing knowledge of the mTOR signaling network. Semin Cell Dev Biol 36:79–90. doi:10.1016/j.semcdb.2014.09.011
Kerestes M, Faigl V, Kulcsar M, Balogh O, Foldi J, Febel H, Chilliard Y, Huszenicza G (2009) Periparturient insulin secretion and whole-body insulin responsiveness in dairy cows showing various forms of ketone pattern with or without puerperal metritis. Domest Anim Endocrinol 37(4):250–261. doi:10.1016/j.domaniend.2009.07.003
Kokkonen T, Taponen J, Anttila T, Syrjala-Qvist L, Delavaud C, Chilliard Y, Tuori M, Tesfa AT (2005) Effect of body fatness and glucogenic supplement on lipid and protein mobilization and plasma leptin in dairy cows. J Dairy Sci 88(3):1127–1141. doi:10.3168/jds.S0022-0302(05)72779-X
Kuhla B, Nurnberg G, Albrecht D, Gors S, Hammon HM, Metges CC (2011) Involvement of skeletal muscle protein, glycogen, and fat metabolism in the adaptation on early lactation of dairy cows. J Proteome Res 10(9):4252–4262. doi:10.1021/pr200425h
Liu W, Thomas SG, Asa SL, Gonzalez-Cadavid N, Bhasin S, Ezzat S (2003) Myostatin is a skeletal muscle target of growth hormone anabolic action. J Clin Endocrinol Metab 88(11):5490–5496. doi:10.1210/jc.2003-030497
Lucy MC, Escalante RC, Keisler DH, Lamberson WR, Mathew DJ (2013) Short communication: glucose infusion into early postpartum cows defines an upper physiological set point for blood glucose and causes rapid and reversible changes in blood hormones and metabolites. J Dairy Sci 96(9):5762–5768. doi:10.3168/jds.2013-6794
Manickam R, Pena RN, Whitelaw CB (2008) Mammary gland differentiation inversely correlates with GDF-8 expression. Mol Reprod Dev 75(12):1783–1788. doi:10.1002/mrd.20918
Mann S, Yepes FA, Overton TR, Wakshlag JJ, Lock AL, Ryan CM, Nydam DV (2015) Dry period plane of energy: effects on feed intake, energy balance, milk production, and composition in transition dairy cows. J Dairy Sci 98(5):3366–3382. doi:10.3168/jds.2014-9024
Mann S, Leal Yepes FA, Duplessis M, Wakshlag JJ, Overton TR, Cummings BP, Nydam DV (2016) Dry period plane of energy: effects on glucose tolerance in transition dairy cows. J Dairy Sci 99(1):701–717
Medina R, Wing SS, Goldberg AL (1995) Increase in levels of polyubiquitin and proteasome mRNA in skeletal muscle during starvation and denervation atrophy. Biochem J 307(Pt 3):631–637
Meijer GA, Van der Meulen J, Bakker JG, Van der Koelen CJ, Van Vuuren AM (1995) Free amino acids in plasma and muscle of high yielding dairy cows in early lactation. J Dairy Sci 78(5):1131–1141. doi:10.3168/jds.S0022-0302(95)76730-3
Oldham JM, Osepchook CC, Jeanplong F, Falconer SJ, Matthews KG, Conaglen JV, Gerrard DF, Smith HK, Wilkins RJ, Bass JJ, McMahon CD (2009) The decrease in mature myostatin protein in male skeletal muscle is developmentally regulated by growth hormone. J Physiol 587(Pt 3):669–677. doi:10.1113/jphysiol.2008.161521
Pasiakos SM, Vislocky LM, Carbone JW, Altieri N, Konopelski K, Freake HC, Anderson JM, Ferrando AA, Wolfe RR, Rodriguez NR (2010) Acute energy deprivation affects skeletal muscle protein synthesis and associated intracellular signaling proteins in physically active adults. J Nutr 140(4):745–751. doi:10.3945/jn.109.118372
Powell SR, Davies KJ, Divald A (2007) Optimal determination of heart tissue 26S-proteasome activity requires maximal stimulating ATP concentrations. J Mol Cell Cardiol 42(1):265–269. doi:10.1016/j.yjmcc.2006.10.010
Roskoski R Jr (2012) ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res 66(2):105–143. doi:10.1016/j.phrs.2012.04.005
Sandri M (2008) Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 23:160–170. doi:10.1152/physiol.00041.2007
Solomon AM, Bouloux PM (2006) Modifying muscle mass - the endocrine perspective. J Endocrinol 191(2):349–360. doi:10.1677/joe.1.06837
Stipanuk MH (2009) Macroautophagy and its role in nutrient homeostasis. Nutr Rev 67(12):677–689. doi:10.1111/j.1753-4887.2009.00252.x
Taniguchi CM, Emanuelli B, Kahn CR (2006) Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol 7(2):85–96. doi:10.1038/nrm1837
Tesseraud S, Grizard J, Debras E, Papet I, Bonnet Y, Bayle G, Champredon C (1993) Leucine metabolism in lactating and dry goats: effect of insulin and substrate availability. Am J Physiol 265(3 Pt 1):E402–E413
van der Drift SG, Houweling M, Schonewille JT, Tielens AG, Jorritsma R (2012) Protein and fat mobilization and associations with serum beta-hydroxybutyrate concentrations in dairy cows. J Dairy Sci 95(9):4911–4920. doi:10.3168/jds.2011-4771
Velloso CP (2008) Regulation of muscle mass by growth hormone and IGF-I. Br J Pharmacol 154(3):557–568. doi:10.1038/bjp.2008.153
Acknowledgments
The authors thank Charlene Ryan, Bryant Stuttle (Cornell University, Ithaca, NY) and the staff of the Cornell University Animal Science research facility in Harford, NY. We acknowledge the help of Mélissa Dupléssis (Université Laval, Québec, Canada) with sample collection.
Funding
This study was supported by the Agriculture and Food Research Initiative competitive Grant No. 2012-67015-30230 from the United States Department of Agriculture National Institute of Food and Agriculture (Washington, DC). A. Abuelo was funded by the Pedro Barrié de la Maza Foundation (A Coruña, Spain).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have any conflict of interest.
Ethical approval
All procedures were approved by the Cornell University Institutional Animal Care and Use Committee (Protocol No. 2011-0016) and were in accordance with the ethical standards of the institution at which the studies were conducted.
Additional information
Communicated by G. Heldmaier.
Electronic supplementary material
Below is the link to the electronic supplementary material.
360_2016_969_MOESM1_ESM.png
Supplementary Fig. 1 Schematic representing the pathways involved in muscle atrophy investigated in this study. Insulin and IGF-1 bind to receptors of the tyrosine kinase type. Signal transduction occurs via insulin receptor substrate (IRS) and leads to increased phosphorylation of AKT downstream. AKT in turn activates mTOR complex 1 (mTORC1) independently of amino acids entering the cell. Activation of mTOR is measured by an increase in direct substrates of this kinase such as S6-Kinase (S6 K) and translation repressor protein 4E-BP1 (4EBP1). Activation of mTORC1 in turn suppresses macroautophagy, measured as the increase in the ratio of microtubule-associated protein 1 light chain 3 (LC3)-II to LC3-I. Macroautophagy is a bulk degradation process of cell components in lysosomes. AKT activation inhibits forkhead box protein transcription factor (FOXO) by phosphorylation, excluding it from the nucleus. Activated FOXO (in case of reduced phosphorylation of AKT) enters the nucleus and upregulates transcription of E3-ligases atrogin-1 and MuRF-1. Proteins destined for proteasomal degradation are tagged with free ubiquitin in a series of steps involving E3-ligases. Ubiquitinated proteins are degrades in the 26S proteasome to smaller peptides. Extracellular signal-regulated kinase (ERK) is also activated by signal transduction through the tyrosine kinase receptor following a number of different growth factors, mitogens, hormones, and other mitogenic stimuli. (PNG 176 kb)
Rights and permissions
About this article
Cite this article
Mann, S., Abuelo, A., Nydam, D.V. et al. Insulin signaling and skeletal muscle atrophy and autophagy in transition dairy cows either overfed energy or fed a controlled energy diet prepartum. J Comp Physiol B 186, 513–525 (2016). https://doi.org/10.1007/s00360-016-0969-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00360-016-0969-1