Abstract
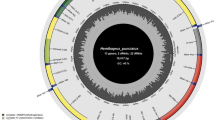
Deep-sea organisms survive in an extremely harsh environment. There must be some genetic adaptation mechanisms for them. We systematically characterized and compared the complete mitochondrial genome (mitogenome) of a deep-sea crab (Chaceon granulates) with those of shallow crabs. The mitogenome of the crab was 16 126 bp in length, and encoded 37 genes as most of a metazoan mitogenome, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and 2 ribosomal RNA (rRNA) genes. The gene arrangement and orientation was conserved in the crabs. However, a unique mitogenome element regulator, the origin of light-strand replication (OL), was firstly predicted in the present crab mitogenome. In addition, further positive selection analysis showed that two residues (33S in ND3 and 502I in ND5) in C. granulates mitogenome were positively selected, indicated the selective evolution of the deep-sea crab. Therefore, the mitogenome of deep-sea C. granulates showed a unique OL element and positive selection. These special features would influence the mitochondrial energy metabolism, and be involved in the adaptation of deep-sea environment, such as oxygen deficits and low temperatures.
Similar content being viewed by others
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the article and its supplementary files.
References
Bernt M, Braband A, Schierwater B, Stadler P F. 2013. Genetic aspects of mitochondrial genome evolution. Molecular Phylogenetics and Evolution, 69(2): 328–338.
Boore J L. 1999. Animal mitochondrial genomes. Nucleic Acids Research, 27(8): 1 767–1 780.
Boore J L. 2001. Complete mitochondrial genome sequence of the polychaete annelid Platynereis dumerilii. Molecular Biology and Evolution, 18(7): 1 413–1 416.
Burkenroad M D. 1981. The higher taxonomy and evolution of Decapoda (Crustacea). Transactions of the San Diego Society of Natural History, 19(17): 251–268.
Clayton D A. 1991. Replication and transcription of vertebrate mitochondrial-DNA. Annual Review of Cell Biology, 7(1): 453–478.
Coscia I, Castilho R, Massa-Gallucci A, Sacchi C, Cunha R L, Stefanni S, Helyar S J, Knutsen H, Mariani S. 2018. Genetic homogeneity in the deep-sea grenadier Macrourus berglax across the North Atlantic Ocean. Deep Sea Research Part I: Oceanographic Research Papers, 132: 60–67.
Da Fonseca R R, Johnson W E, O’Brien S J, Ramos M J, Antunes A. 2008. The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics, 9: 119.
Etter R J, Rex M A, Chase M C, Quattro J M. 1999. A genetic dimension to deep-sea biodiversity. Deep Sea Research Part I: Oceanographic Research Papers, 46(6): 1 095–1 099.
Fernández-Silva P, Enriquez J A, Montoya J. 2003. Replication and transcription of mammalian mitochondrial DNA. Experimental Physiology, 88(1): 41–56.
Flot J F, Tillier S. 2007. The mitochondrial genome of Pocillopora (Cnidaria: Scleractinia) contains two variable regions: The putative D-loop and a novel ORF of unknown function. Gene, 401(1–2): 80–87.
Green D R, Reed J C. 1998. Mitochondria and apoptosis. Science, 281(5381): 1 309–1 312.
Gu M L, Dong X Q, Shi L, Shi L, Lin K Q, Huang X Q, Chu J Y. 2012. Differences in mtDNA whole sequence between Tibetan and Han populations suggesting adaptive selection to high altitude. Gene, 496(1): 37–44.
Guo X H, Liu S J, Liu Y. 2003. Comparative analysis of the mitochondrial DNA control region in cyprinids with different ploidy level. Aquaculture, 224(1–4): 25–38.
Hassanin A, Léger N, Deutsch J. 2005. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Systematic Biology, 54(2): 277–298.
Hassanin A, Ropiquet A, Couloux A, Cruaud C. 2009. Evolution of the mitochondrial genome in mammals living at high altitude: new insights from a study of the Tribe Caprini (Bovidae, Antilopinae). Journal of Molecular Evolution, 68(4): 293–310.
Hui M, Cheng J, Sha Z L. 2018. Adaptation to the deep-sea hydrothermal vents and cold seeps: Insights from the transcriptomes of Alvinocaris longirostris in both environments. Deep Sea Research Part I: Oceanographic Research Papers, 135: 23–33.
Jacobsen M W, Da Fonseca R R, Bernatchez L, Hansen M M. 2016. Comparative analysis of complete mitochondrial genomes suggests that relaxed purifying selection is driving high nonsynonymous evolutionary rate of the NADH2 gene in whitefish (Coregonus ssp.). Molecular Phylogenetics and Evolution, 95: 161–170.
Jebbar M, Franzetti B, Girard E, Oger P. 2015. Microbial diversity and adaptation to high hydrostatic pressure in deep-sea hydrothermal vents prokaryotes. Extremophiles, 19(4): 721–740.
Jiang L C, Wang G C, Tan S, Gong S A, Yang M, Peng Q K, Peng R, Zou F D. 2013. The complete mitochondrial genome sequence analysis of Tibetan argali (Ovis ammon hodgsoni): implications of Tibetan argali and Gansu argali as the same subspecies. Gene, 521(1): 24–31.
Jin X X, Wang R X, Wei T, Tang D, Xu T J. 2015. Complete mitochondrial genome sequence of Tridentiger bifasciatus and Tr dent ger barbatus (Perciformes, Gobiidae): a mitogenomic perspective on the phylogenetic relationships of Gobiidae. Molecular BiologyReports, 42(1): 253–265.
Kawaguchi A, Miya M, Nishida M. 2001. Complete mitochondrial DNA sequence of Aulopus japonicus (Teleostei: Aulopiformes), a basal Eurypterygii: longer DNA sequences and higher-level relationships. Ichthyological Research, 48(3): 213–223.
Lavrov D V, Brown W M, Boore J L. 2000. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proceedings of the National Academy of Sciences of the United States of America, 97(25): 13 738–13 742.
Liao F, Wang L, Wu S, Li Y P, Zhao L, Huang G M, Niu C J, Liu Y Q, Li M G. 2010. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). International Journal of Biological Sciences, 6(2): 172–186.
Liao Y Y, Mo G D, Sun J L, Wei F Y, Liao D J. 2016. Genetic diversity of Guangxi chicken breeds assessed with microsatellites and the mitochondrial DNA D-loop region. Molecular Biology Reports, 43(5): 415–425.
Liu Y, Cui Z X. 2010. Complete mitochondrial genome of the Asian paddle crab Charybdis japonica (Crustacea: Decapoda: Portunidae): gene rearrangement of the marine brachyurans and phylogenetic considerations of the decapods. Molecular BiologyReports, 37(5): 2 559–2 569.
Lowe T M, Eddy S R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research, 25(5): 955–964.
Luo Y J, Gao W X, Gao Y Q, Tang S, Huang Q Y, Tan X L, Chen J, Huang T S. 2008. Mitochondrial genome analysis of Ochotona curzoniae and implication of cytochrome c oxidase in hypoxic adaptation. Mitochondrion, 8(5–6): 352–357.
Ma H Y, Ma C Y, Li C H, Lu J X, Zou X, Gong Y Y, Wang W, Chen W, Ma L B, Xia L J. 2015. First mitochondrial genome for the red crab (Charybdis feriata) with implication of phylogenomics and population genetics. Scientific Reports, 5: 11 524.
Ma H Y, Ma C Y, Li X C, Xu Z, Feng N N, Ma L B. 2013. The complete mitochondrial genome sequence and gene organization of the mud crab (Scylla paramamosain) with phylogenetic consideration. Gene, 519(1): 120–127.
Marshall H D, Baker A J. 1997. Structural conservation and variation in the mitochondrial control region of fringilline finches (Fringilla spp.) and the greenfinch (Carduelis chloris). Molecular Biology and Evolution, 14(2): 173–184.
Miller A D, Murphy N P, Burridge C P, Austin C M. 2005. Complete mitochondrial DNA sequences of the decapod crustaceans Pseudocarcinus gigas (Menippidae) and Macrobrachium rosenbergii (Palaemonidae). Marine Biotechnology, 7(4): 339–349.
Newmeyer D D, Ferguson-Miller S. 2003. Mitochondria: releasing power for life and unleashing the machineries of death. Cell, 112(4): 481–490.
Ning T, Xiao H, Li J, Hua S, Zhang Y P. 2010. Adaptive evolution of the mitochondrial ND6 gene in the domestic horse. Genetics and Molecular Research, 9(1): 144–150.
Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature, 290(5806): 470–474.
Perna N T, Kocher T D. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. Journal of Molecular Evolution, 41(3): 353–358.
Posada D, Crandall K A. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics, 14(9): 817–818.
Rex M A. 1981. Community structure in the deep-sea Benthos. Annual Review of Ecology and Systematics, 12(1): 331–353.
Sanders H L, Hessler R R. 1969. Ecology of the deep-sea benthos. Science, 163(3874): 1 419–1 424.
Sbisa E, Tanzariello F, Reyes A, Pesole G, Saccone C. 1997. Mammalian mitochondrial D-loop region structural analysis: identification of new conserved sequences and their functional and evolutionary implications. Gene, 205(1–2): 125–140.
Shen X, Ren J F, Cui Z X, Sha Z L, Wang B, Xiang J H, Liu B. 2007. The complete mitochondrial genomes of two common shrimps (Litopenaeus vannamei and Fenneropenaeus chinensis) and their phylogenomic considerations. Gene, 403(1–2): 98–109.
Sogin M L, Morrison H G, Huber J A, Welch D M, Huse S M, Neal P R, Arrieta J M, Herndl G J. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proceedings of the National Academy of Sciences of the United States of America, 103(32): 12 115–12 120.
Sun S E, Hui M, Wang M X, Sha Z L. 2018. The complete mitochondrial genome of the alvinocaridid shrimp Shinkaicaris leurokolos (Decapoda, Caridea): insight into the mitochondrial genetic basis of deep-sea hydrothermal vent adaptation in the shrimp. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 25: 42–52.
Tang B P, Liu Y, Xin Z Z, Zhang D Z, Wang Z F, Zhu X Y, Wang Y, Zhang H B, Zhou C L, Chai X Y, Liu Q N. 2017. Characterisation of the complete mitochondrial genome of Helice wuana (Grapsoidea: Varunidae) and comparison with other Brachyuran crabs. Genomics, 110(4): 221–230.
Tsang L M, Ma K Y, Ahyong S T, Chan T Y, Chu K H. 2008. Phylogeny of Decapoda using two nuclear protein-coding genes: origin and evolution of the Reptantia. Molecular Phylogenetics and Evolution, 48(1): 359–368.
Umetsu K, Iwabuchi N, Yuasa I, Saitou N, Clark P F, Boxshall G, Osawa M, Igarashi K. 2002. Complete mitochondrial DNA sequence of a tadpole shrimp (Triops cancriformis) and analysis of museum samples. Electrophoresis, 23(24): 4 080–4 084.
Valverde J R, Batuecas B, Moratilla C, Marco R, Garesse R. 1994. The complete mitochondrial DNA sequence of the crustacean Artemia franciscana. Journal of Molecular Evolution, 39: (4).
Wang Z L, Li C, Fang W Y, Yu X P. 2016. The complete mitochondrial genome of two Tetragnatha spiders (Araneae: Tetragnathidae): severe truncation of tRNAs and novel gene rearrangements in Araneae. International Journal of Biological Sciences, 12(1): 109–119.
Yamauchi M M, Miya M U, Nishida M. 2003. Complete mitochondrial DNA sequence of the swimming crab, Portunus trituberculatus (Crustacea: Decapoda: Brachyura). Gene, 311: 129–135.
Yu L, Wang X P, Ting N, Zhang Y P. 2011. Mitogenomic analysis of Chinese snub-nosed monkeys: evidence of positive selection in NADH dehydrogenase genes in high-altitude adaptation. Mitochondrion, 11(3): 497–503.
Zhang B, Zhang Y H, Wang X, Zhang H X, Lin Q. 2017a. The mitochondrial genome of a sea anemone Bolocera sp. exhibits novel genetic structures potentially involved in adaptation to the deep-sea environment. Ecology and Evolution, 7(13): 4 951–4 962.
Zhang D X, Szymura J M, Hewitt G M. 1995. Evolution and structural conservation of the control region of insect mitochondrial DNA. Journal of Molecular Evolution, 40(4): 382–391.
Zhang Q L, Zhang L, Zhao T X, Wang J, Zhu Q H, Chen J Y, Yuan M L. 2017b. Gene sequence variations and expression patterns of mitochondrial genes are associated with the adaptive evolution of two Gynaephora species (Lepidoptera: Lymantriinae) living in different high-elevation environments. Gene, 610: 148–155.
Zhang Y, Li X G, Bartlett D H, Xiao X. 2015. Current developments in marine microbiology: high-pressure biotechnology and the genetic engineering of piezophiles. Current Opinion in Biotechnology, 33: 157–164.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the Strategic Priority Research Program of Chinese Academy of Sciences (Nos. XDA13020103, XDA19060301), the National Science & Technology Fundamental Resources Investigation Program of China (No. 2018FY100106), and the National Key Research and Development Program “Marine Environment Security Project” (Nos. 2017YFC0506302, 2018YFC1406503)
Electronic supplementary material
343_2019_8364_MOESM1_ESM.pdf
Comparative analysis of mitochondrial genome of a deep-sea crab Chaceon granulates reveals positive selection and novel genetic features
Rights and permissions
About this article
Cite this article
Zhang, B., Wu, Y., Wang, X. et al. Comparative analysis of mitochondrial genome of a deep-sea crab Chaceon granulates reveals positive selection and novel genetic features. J. Ocean. Limnol. 38, 427–437 (2020). https://doi.org/10.1007/s00343-019-8364-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00343-019-8364-x