Abstract
Comparative sequence analyses have identified highly conserved genomic DNA sequences, including noncoding sequences, between humans and other species. By performing whole-genome comparisons of human and mouse, we have identified 611 conserved noncoding sequences longer than 500 bp, with more than 95% identity between the species. These long conserved noncoding sequences (LCNS) include 473 new sequences that do not overlap with previously reported ultraconserved elements (UCE), which are defined as aligned sequences longer than 200 bp with 100% identity in human, mouse, and rat. The LCNS were distributed throughout the genome except for the Y chromosome and often occurred in clusters within regions with a low density of coding genes. Many of the LCNS were also highly conserved in other mammals, chickens, frogs, and fish; however, we were unable to find orthologous sequences in the genomes of invertebrate species. In order to examine whether these conserved sequences are functionally important or merely mutational cold spots, we directly measured the frequencies of ENU-induced germline mutations in the LCNS of the mouse. By screening about 40.7 Mb, we found 35 mutations, including mutations at nucleotides that were conserved between human and fish. The mutation frequencies were equivalent to those found in other genomic regions, including coding sequences and introns, suggesting that the LCNS are not mutational cold spots at all. Taken together, these results suggest that mutations occur with equal frequency in LCNS but are eliminated by natural selection during the course of evolution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Comparative genomics has revealed that a large number of noncoding DNA sequences are conserved between humans and other species. However, there is little information about the functional roles of these conserved noncoding sequences (CNS), which, surprisingly, are often much more highly conserved than nucleotide sequences encoding well-conserved proteins. Comparisons of genomic sequences among various vertebrate species have revealed many CNS, which are known by various names (Ahituv et al. 2005; Bejerano et al. 2004; Dermitzakis et al. 2002; Margulies et al. 2003; Persampieri et al. 2008; Prabhakar et al. 2006; Sandelin et al. 2004; Shin et al. 2005; Siepel et al. 2005; Thomas et al. 2003; Venkatesh et al. 2006; Visel et al. 2008; Woolfe et al. 2005). For instance, 2262 CNS (conserved nongenic; length ≥100 bp and identity ≥70%) were found by comparing human chromosome 21 and the syntenic mouse region (Dermitzakis et al. 2002). Nearly 5000 CNS have been found in comparisons between human and fish (Sandelin et al. 2004; Shin et al. 2005; Venkatesh et al. 2006; Woolfe et al. 2005). In addition, Bejerano et al. (2004) have identified 481 ultraconserved elements (UCE) of more than 200 bp with 100% identity among the human, mouse, and rat genomes. The definition of UCE is not restricted to noncoding sequences, so UCE can include coding sequences as well as CNS.
Several studies have suggested that these conserved sequences transcriptionally regulate developmental genes. Indeed, some CNS have tissue-specific enhancer activity (Bailey et al. 2006; Nobrega et al. 2003; Pennacchio et al. 2006; Prabhakar et al. 2006; Shin et al. 2005; Visel et al. 2008; Woolfe et al. 2005). CNS have also been associated with the long-range regulation of gene expression (Loots et al. 2000; Nobrega et al. 2003; Sabherwal et al. 2007; Sagai et al. 2005). Some studies have even provided genetic evidence that CNS have biological functions; for example, point mutations in a CNS are responsible for mouse and human preaxial polydactyly with mirror-image digit duplications (Masuya et al. 2007; Sagai et al. 2004). On the other hand, deleting megabases of the mouse genome, including many CNS, did not induce an abnormal phenotype (Nobrega et al. 2004). Therefore, additional studies are needed to determine whether CNS are generally functional.
Two hypotheses are proposed to explain the high conservation of CNS. One hypothesis is that they are selectively constrained, and the other hypothesis is that CNS are merely mutational cold spots. A recent analysis of genotype data in human SNP projects implied that CNS are not mutational cold spots (Drake et al. 2006; Katzman et al. 2007). However, the hypothesis that CNS are mutational cold spots, regardless of their functional importance, has not been experimentally examined.
To directly examine whether CNS are mutational cold spots, we have identified a new class of CNS that we call long conserved noncoding sequences (LCNS). We examined the frequency and positions of LCNS and UCE in the mouse genome and investigated the conservation of these elements across species. We also studied LCNS mutation rates in the mouse and have excluded the “cold spot” hypothesis by directly assessing the mutation frequency of N-ethyl-N-nitrosourea (ENU)-induced substitutions in CNS.
Materials and methods
Extraction of LCNS: sequence data and alignment
We compared and extracted conserved noncoding sequences from human and mouse genomes three times between 2002 and 2007 using the latest data set at each time point. A total of 611 sequences were extracted.
First extraction To compare human and mouse genomic sequences, whole genomic sequences of Golden Path (repeat masked) in human build 34 (hg16) and mouse build 32 (mm4), which had masked repetitive regions as “N,” were retrieved from the UCSC genome browser (http://genome.ucsc.edu/). The genomic sequences were aligned using BLAST. To exclude coding sequences, the resulting fragments were searched with “mrna.fa,” which is a data set of mRNAs from the selected species in GenBank. Matching fragments were removed. We identified 444 sequences longer than 500 bp with more than 95% identity between human and mouse. The number of LCNS was reduced to 411 after several updates of the genomic sequence data, whose latest versions were hg18 and mm9.
Second extraction Whole genomic sequences of Golden Path (repeat masked) in human build 36 (hg18) and mouse build 35 (mm7) were retrieved from the UCSC genome browser. We masked all of the coding sequences in the human and mouse genomic sequences as “N,” referring to Ensembl information (www.ensembl.org) to identify genes, transcripts, exons, and coding sequences. By aligning the masked genomes using BLAST, we obtained 508 LCNS (≥500 bp and ≥95% identity). We used TSUBAME (Tokyo-Tech Supercomputer and Ubiquitously Accessible Mass-Storage Environment), which is a supergrid computer cluster at the Tokyo Institute of Technology, to search these sequences with BLAST. Of the 508 sequences, 298 were identical to those from the first extraction. Thus, 210 new sequences were extracted. After renewal of the database (from mm7 to mm8) and detailed inspection of conformation to the definition of LCNS, the number for the newly extracted sequences was 194.
Third extraction We obtained a new data set of mouse genomic sequence (build 36, mm8) and extracted LCNS by almost the same method as the second extraction. Six new sequences were extracted. We used RSCC (Riken Super Combined Cluster system) instead of TSUBAME for this extraction.
The location information of the identified sequences in the human and mouse genomes are listed in Supplementary Table 1, which includes the links to the genomic information on the UCSC genome browser (http://genome.ucsc.edu). It provides the actual nucleotide sequences and additional information about each sequence. The information is based on human build 36 (hg18) and mouse build 37 (mm9).
Annotation of LCNS
The information about the nearest-neighboring coding genes was obtained from the Ensembl database. The position and other information for each coding gene were obtained from BioMart or Application Program Interface (API) in Perl.
For comparing LCNS among multiple species, whole genomic sequences of dog (Canis familiaris), chicken (Gallus gallus), frog (Xenopus tropicalis), fugu (Takifugu rubripes), tetraodon (Tetraodon nigroviridis), zebrafish (Danio rerio), two Ascidiacea species (Ciona intestinalis and Ciona savignyi), and fruit fly (Drosophila melanogaster) were obtained from the UCSC genome browser or Ensembl website. BLAST searches of each genomic sequence were conducted using the 611 mouse LCNS as queries.
Measurement of the mutation frequency
We used the RIKEN mutant mouse library to measure the frequency of ENU-induced mutations using temperature-gradient capillary electrophoresis as described previously (Sakuraba et al. 2005). Primers used in the screening are listed in Supplementary Table 3. The 35 mutant mouse lines obtained in this analysis are available from RIKEN BioResource Center (http://www.brc.riken.jp/lab/mutants/genedriven.htm).
Comparison of LCNS and visualization
The VISTA program (Frazer et al. 2004a; Mayor et al. 2000) was used to compare LCNS among human, mouse, chicken, frog, and zebrafish. We used a 100-bp window and 70% conservation level for mouse–human, mouse–chicken, mouse–frog, and mouse–zebrafish comparisons.
Results
Identification of LCNS
We compared whole genomic human and mouse sequences by BLAST searching and then extracted CNS using the parameters of ≥95% identity and ≥500 bp in length. As described in the Materials and methods section, we searched for CNS three times in different versions of the database since 2002. We identified a total of 611 long conserved noncoding sequences (LCNS; Supplementary Table 1). To check for redundancy among the 611 LCNS, we examined the similarities between all the sequences with a self-BLAST search. Six pairs of 12 sequences were found to be highly homologous (Supplementary Table 2). The remaining 599 sequences were unique and no obvious consensus sequences were found.
Distribution and locations of the LCNS
The LCNS were distributed among all of the chromosomes except for the Y chromosome in both the human and mouse genomes (Supplementary Table 1, Figs. 1 and 2). However, the numbers of LCNS on each chromosome varied and were not proportional to the length of the LCNS extractable sequences (noncoding and nonrepetitive sequences; Fig. 2). In addition to the interchromosomal bias, the intrachromosomal distributions of LCNS were uneven as well. Mouse chromosome 7 was a typical case, with the LCNS concentrated in several areas of the chromosome rather than distributed randomly (Fig. 1), indicating that many LCNS exist as clusters.
Number of LCNS on each chromosome. The size of LCNS extractable genomic sequences (noncoding and nonrepetitive sequences) and number of LCNS extracted from human (a) and mouse (b) chromosomes. The total bp of coding and repetitive sequences (upper white bars) and noncoding and nonrepetitive sequences (lower gray bars) are shown for each chromosome. The total bp (left axis) represents the length of each chromosome. The numbers of LCNS on each chromosome are indicated by the black dots and numbers on the right axis. The number of LCNS per 100 Mb of LCNS extractable area in human (c) and mouse (d) chromosomes. The horizontal dotted lines represent the average values (Avg)
Based on the information in the Ensembl mouse genome database, we classified each LCNS as “intronic,” “intergenic,” or “untranslated region (UTR)” (Supplementary Table 1). About 55% of LCNS were located in intergenic regions and 41% were within introns. Only 4% of LCNS were in UTRs.
Comparison of LCNS with UCE
The extraction parameters for the LCNS (≥95% and ≥500 bp) were extremely stringent, which is very comparable to those for the UCE [100% and ≥200 bp (Bejerano et al. 2004)]. This was indicated by the fact that similar numbers of LCNS (611) and UCE (481) were extracted. Although the extraction stringencies were equivalent, the characteristics of the LCNS were quite different from those of the UCE. Unlike the LCNS, which are extracted from noncoding and nonrepetitive sequences, UCE do not exclude coding sequences. Therefore, 69 and 9 UCE overlapping coding sequences and repetitive sequences, respectively, were subtracted from the sequence comparison, for a total of 403 UCE and 611 LCNS. We first examined whether the individual sequences of LCNS overlapped those of UCE. One hundred fifty (37%) of the 403 UCE overlapped with 138 (23%) of the 611 LCNS. By definition, LCNS are usually larger than UCE, and 12 LCNS included 2 different UCE. The remaining 63% of the UCE and 77% of the LCNS were unique in the data sets. We have therefore identified 473 new highly conserved LCNS sequences that do not overlap with UCE.
We examined the positional relationships of the 611 LCNS and the 472 nonrepetitive UCE, excluding the 9 UCE overlapping repetitive sequences from the 481 total UCE. Both LCNS and UCE were scattered all over the genome, with some forming clusters (Supplementary Fig. 1). Each cluster was composed of both LCNS and UCE, indicating that the distribution profiles of the LCNS and UCE were similar at this resolution. Next, we examined the neighboring coding genes for both UCE and LCNS. Of the 472 UCE, the neighboring genes of 435 were identical to the neighboring genes of LCNS; the neighboring genes of 513 of the 611 LCNS were identical to those of UCE. These results suggest that highly conserved sequences, such as LCNS and UCE, are concentrated in the same intergenic regions or in the introns of the same gene and that these regions are distributed throughout the genome.
LCNS tend to be in regions of low gene density
For both intronic and intergenic LCNS, the distance to the nearest coding exon was often very long. Of the 611 LCNS, 402 were 10 kb or more from the nearest coding sequences. Moreover, 150 LCNS were 100 kb or more away and 4 were 1 Mb or more away (23 were ≥500 kb away). Interestingly, despite the long distances, the genes nearest to a LCNS were usually the same in human and mouse and were oriented in the same direction, indicating their long syntenic conservation. We determined the number of coding genes within ± 1 Mb of LCNS or genes. Although there was an average of 30.0 genes within 1 Mb of a given gene, there was an average of only 10.0 genes within 1 Mb of a LCNS. These results suggest that, like the UCE, LCNS tend to exist in regions with a low density of coding genes.
Conservation in other species
We examined the conservation of LCNS in various vertebrates and invertebrates. Using the 611 human-mouse LCNS as queries, we searched the genomic databases of nine species (dog, chicken, frog, fugu, tetraodon, zebrafish, two Ascidiacea [Ciona intestinalis, Ciona savignyi], and fruit fly) by BLAST analysis (e-value = 1e-50, ≥100 bp; Table 1). Almost all of the LCNS (606/611) were also conserved in the dog. Chicken and frog had 81% (493/611) and 65% (397/611) of the LCNS, respectively. The three fish species had 9–14% (58-83 of 611) of the LCNS. However, the searches found no LCNS in the two Ascidiacea species or fruit fly. These results indicate that the LCNS that are common to human and mouse exist widely in vertebrates but not in invertebrates.
Mutation frequency
To examine whether LCNS are mutational cold spots, we compared the mutation frequencies in LCNS with other genomic regions. For this purpose, we measured the frequency of ENU-induced germline mutations in mice. We used the RIKEN mutant mouse library, a collection of genomic DNA from F1 progeny (G1) of ENU-mutagenized C57BL/6 J males and untreated females (Sakuraba et al. 2005). Because ENU-induced mutations are heterozygous in the G1 mice, all mutations, except for dominant lethal mutations, can be detected by sequence-based screening of the RIKEN library. In our previous study, we found 148 ENU-induced mutations in a 197-Mb screening (Table 2a), for an overall mutation frequency of 1 per 1.33 Mb (Sakuraba et al. 2005). In this experiment, we found 12 mutations in a 16.4-Mb screening of nine randomly chosen LCNS (Table 2b), for a mutation frequency of 1 per 1.37 Mb, which is equivalent to that of other genomic regions, including coding sequences and introns.
After we published the previous report (Sakuraba et al. 2005), we improved our screening method by using a high-resolution gel system to increase the mutation detection rate. We found 230 new mutations from an extensive screening of 248 Mb, including 48 genes and 7 LCNS, for a mutation frequency of 1 per 1.08 Mb (Table 3a). Using this new system, we found 23 mutations from a 24.2-Mb screening of 7 LCNS (Table 3b), including 3 amplicons from our previous report (Sakuraba et al. 2005) and 4 new amplicons. The mutation frequency from the LCNS screening was 1 per 1.05 Mb, which was equivalent to that from the total screening even in two independent screens.
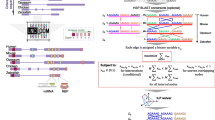
We thus examined the mutation frequency of a total of 12 LCNS in two analyses using different gel systems and found no difference in the frequencies between the LCNS and the other genomic regions. As shown in Fig. 3, we found mutations even at nucleotides that were conserved between human and zebrafish. These results indicate that ENU-induced mutations were equally likely to occur in LCNS, and therefore LCNS are not mutational cold spots. Five of the 12 LCNS in this experiment overlapped with 6 UCE (Tables 2b and 3b), and 9 of 35 LCNS mutations were found in sequences that overlapped between LCNS and UCE. This suggests that like LCNS, UCE are also not mutational cold spots.
Examples of mutations found in LCNS. A typical LCNS and its mutations. (a) LCNS ID 418. The conservation levels among multiple species are presented as a VISTA graph. Gray bars represent two UCE within the LCNS. Vertical blue arrows indicate nucleotide substitution sites due to ENU mutagenesis. Horizontal arrows indicate primer pairs used in the mutation screening. (b) Sequencing chromatograms of the mutation sites are shown in the upper part of each panel and sequence alignments of the mutation sites in multiple species are shown in the lower part of each panel. The upper and lower chromatograms are reference and mutant sequences, respectively. Arrowheads indicate the mutation sites. “K,” “W,” and “R” indicate G/T, A/T, and A/G, respectively, based on IUB code
Discussion
We have identified 611 noncoding sequences that are longer than 500 bp and have more than 95% identity between the human and mouse genomes. These LCNS are distributed throughout the genome except for the Y chromosome. Similar to other CNS, LCNS have several interesting characteristics: (1) They form clusters and are concentrated in specific genomic regions. (2) They tend to be located far from coding sequences. Even intronic LCNS are often separated from neighboring coding exons by more than 10 kb. As yet, we cannot explain why they are separated from coding sequences, but the distance may be important for their mechanism of action, such as long-range regulation of gene expression (Kleinjan and van Heyningen 2005; Loots et al. 2000, 2005; Masuya et al. 2007; Nobrega et al. 2003; Sabherwal et al. 2007). (3) In addition to sequence conservation, the distances and orientations between LCNS and neighboring coding sequences (genes) are also conserved among multiple species, i.e., the syntenic relationship is conserved. These characteristics of LCNS are consistent with previous observations of CNS (Bejerano et al. 2004; de la Calle-Mustienes et al. 2005; Dermitzakis et al. 2002; Margulies et al. 2003; Sandelin et al. 2004; Shin et al. 2005; Thomas et al. 2003; Venkatesh et al. 2006; Woolfe et al. 2005). We have extracted the LCNS as a very small fraction of the CNS using extremely stringent conditions. Thus, potentially, the nature of the LCNS could be quite different from the general characteristics of CNS; or at least LCNS could consist of a very biased fraction of CNS. However, the above-mentioned similar characteristics between LCNS and CNS indicate that the LCNS are not an extreme fraction of CNS; rather, we consider that the LCNS are very typical members of CNS. Therefore, the LCNS should provide a general resource for the functional studies of CNS. It is not practical to conduct functional studies on thousands of CNS one by one; however, it is very feasible to experimentally examine the function of 611 LCNS and/or 481 UCE (Bejerano et al. 2006; Chen et al. 2007; Derti et al. 2006; Gardiner et al. 2006).
We found sequences orthologous to human-mouse LCNS in this study, not only in chicken and frog, but also in fish. However, we did not find these sequences in the invertebrates Ascidiacea and fruit fly. Woolfe et al. (2005) have identified 1400 highly conserved noncoding sequences through sequence comparisons between human and fugu, but they also did not find any similar sequences in invertebrate genomes. These results suggest that the functions of CNS identified by sequence comparisons among vertebrate species may be specific to vertebrates. Although no orthologous sequences of vertebrate CNS have been found in invertebrates, there are independent sets of CNS, not only in insects, but also in nematode, yeast, and plant genomes (Glazov et al. 2005; Guo and Moose 2003; Inada et al. 2003; Siepel et al. 2005). Furthermore, the categories of genes neighboring insect CNS are similar to those near vertebrate CNS (Glazov et al. 2005). The most common feature of eukaryotic CNS, including those from vertebrates, invertebrates, and plants, is their abundance near genes encoding transcriptional factors. Thus, the regulation of gene expression is a universal candidate for CNS function. In vertebrates, several experiments have shown that a portion of CNS actually have enhancer activity (Frazer et al. 2004b), particularly tissue-specific enhancer activity (Bailey et al. 2006; Nobrega et al. 2003; Pennacchio et al. 2006; Prabhakar et al. 2006; Shin et al. 2005; Visel et al. 2008; Woolfe et al. 2005).
Recently, it was shown that some UCE are associated with alternative splicing coupled with nonsense-mediated decay (Lareau et al. 2007; Ni et al. 2007), and Choi et al. (2006) have shown that tissue-specific transcription factors generally have the greatest conservation in their noncoding regions. These data suggest that CNS are associated with strict spatial and temporal regulation of gene expression. However, the mechanisms of the regulation associated with UCE remain to be elucidated. CNS are likely to have various biological functions in addition to transcriptional regulation. Further genetic and molecular analyses of CNS will be needed to reveal the functions and mechanisms.
In general, we expect that nucleotide sequences have been conserved as a result of natural selection during evolution and that the conserved sequences are biologically important. Several previous studies have suggested that the conservation of CNS is due to purifying selection and that CNS are likely to be functional (Keightley et al. 2005; Kryukov et al. 2005). However, a mechanism might exist to protect specific DNA sequences from mutations, leading to conservation of the sequences. In this case, two possibilities may be considered. One is that the DNA within CNS is more strongly protected from mutagens than the DNA in other genomic regions, and the other is that DNA damage in CNS is more likely to be repaired than in other regions. Although there is no evidence for either possibility, the hypothesis that such conserved sequences are mutational cold spots had not previously been ruled out. In this study, we observed ENU-induced mutations in both LCNS and UCE (Fig. 3, Tables 2b and 3b). We found a total of 35 mutations in LCNS from a 40.7-Mb mutation screening, a mutation frequency equivalent to that in other genomic regions (Tables 2a, 2b and 3a, 3b). This result indicates that LCNS are not mutational cold spots and that mutations appear to have occurred equally in LCNS and other regions during evolution. It would be ideal to measure the spontaneous mutation rate in LCNS with the same experimental flow; however, it is not practically possible to conduct such experiments. The analysis using ENU mutagenesis is one of the best assessments to evaluate the susceptibility of whole chromatin structures and genomic DNA sequences against any mutagenic agents. Our direct experimental evidence is consistent with the results of human SNP analyses, which have indirectly implied that these CNS are not mutational cold spots (Drake et al. 2006; Katzman et al. 2007). Taking this information together, we propose that, in general, CNS, LCNS, and UCE are highly conserved not because they are mutational cold spots but because of functional constraints during evolution.
Our next objective will be to investigate the biological functions of CNS using genetic analysis of CNS mutants. However, it might be difficult to detect phenotypic differences between wild types and mutants by general laboratory experiments, because mutations in these conserved sequences might be only slightly deleterious despite the high degree of conservation (Chen et al. 2007; Keightley et al. 2005; Kryukov et al. 2005). Indeed, large deletions of genomic sequences containing many CNS did not affect the mouse phenotype (Nobrega et al. 2004). In addition, some lines of mice lacking UCE failed to reveal any critical abnormalities (Ahituv et al. 2007). On the other hand, several lines of genetic evidence have indicated that deletions of CNS can lead to specific phenotypes. For example, patients with Leri-Weill dyschondrosteosis have an intact SHOX coding gene, but a region located downstream of the gene, including the CNS, is deleted (Sabherwal et al. 2007). A patient with Van Buchem disease has a deletion of a large noncoding region, including seven CNS, located downstream of the SOST coding gene (Loots et al. 2005). The deletion of a conserved noncoding region in intron 5 of the Lmbr1 locus, 1 Mb away from the sonic hedgehog (Shh) coding sequence, resulted in a complete loss of Shh expression in the limb bud and degeneration of skeletal elements distal to the stylopod/zygopod junction (Sagai et al. 2005). In addition, point mutations in this region affect Shh expression and are responsible for mouse and human preaxial polydactyly (Lettice et al. 2002, 2003; Sagai et al. 2004). These results suggest that in addition to mouse deletion mutants, mouse point mutations could be useful for functional analyses of CNS. All 35 of the ENU-induced germline mutations that we identified (Tables 2b and 3b) are preserved in frozen sperm, which can be used to reproduce the mice with these mutations (Sakuraba et al. 2005). These mutant lines are available from the RIKEN BioResource Center. Using this RIKEN mutant mouse library, we have already shown that the gene-driven system for ENU-induced mutations is an effective approach for exploring the functions of CNS and potential cis-regulatory elements (Masuya et al. 2007). We hope that genetic analyses using this resource will reveal the functions of CNS and the mechanisms of their conservation.
References
Ahituv N, Prabhakar S, Poulin F, Rubin EM, Couronne O (2005) Mapping cis-regulatory domains in the human genome using multi-species conservation of synteny. Hum Mol Genet 14:3057–3063
Ahituv N, Zhu Y, Visel A, Holt A, Afzal V, Pennacchio LA, Rubin EM (2007) Deletion of ultraconserved elements yields viable mice. PLoS Biol 5:e234
Bailey PJ, Klos JM, Andersson E, Karlen M, Kallstrom M, Ponjavic J, Muhr J, Lenhard B, Sandelin A, Ericson J (2006) A global genomic transcriptional code associated with CNS-expressed genes. Exp Cell Res 312:3108–3119
Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D (2004) Ultraconserved elements in the human genome. Science 304:1321–1325
Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D (2006) A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441:87–90
Chen CT, Wang JC, Cohen BA (2007) The strength of selection on ultraconserved elements in the human genome. Am J Hum Genet 80:692–704
Choi SS, Bush EC, Lahn BT (2006) Different classes of tissue-specific genes show different levels of noncoding conservation. Genomics 87:433–436
de la Calle-Mustienes E, Feijóo CG, Manzanares M, Tena JJ, Rodríguez-Seguel E, Letizia A, Allende ML, Gómez-Skarmeta JL (2005) A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res 15:1061–1072
Dermitzakis ET, Reymond A, Lyle R, Scamuffa N, Ucla C, Deutsch S, Stevenson BJ, Flegel V, Bucher P, Jongeneel CV, Antonarakis SE (2002) Numerous potentially functional but non-genic conserved sequences on human chromosome 21. Nature 420:578–582
Derti A, Roth FP, Church GM, Wu CT (2006) Mammalian ultraconserved elements are strongly depleted among segmental duplications and copy number variants. Nat Genet 38:1216–1220
Drake JA, Bird C, Nemesh J, Thomas DJ, Newton-Cheh C, Reymond A, Excoffier L, Attar H, Antonarakis SE, Dermitzakis ET, Hirschhorn JN (2006) Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet 38:223–227
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004a) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32:W273–W279
Frazer KA, Tao H, Osoegawa K, de Jong PJ, Chen X, Doherty MF, Cox DR (2004b) Noncoding sequences conserved in a limited number of mammals in the SIM2 interval are frequently functional. Genome Res 14:367–372
Gardiner EJ, Hirons L, Hunter CA, Willett P (2006) Genomic data analysis using DNA structure: an analysis of conserved nongenic sequences and ultraconserved elements. J Chem Inf Model 46:753–761
Glazov EA, Pheasant M, McGraw EA, Bejerano G, Mattick JS (2005) Ultraconserved elements in insect genomes: a highly conserved intronic sequence implicated in the control of homothorax mRNA splicing. Genome Res 15:800–808
Guo H, Moose SP (2003) Conserved noncoding sequences among cultivated cereal genomes identify candidate regulatory sequence elements and patterns of promoter evolution. Plant Cell 15:1143–1158
Inada DC, Bashir A, Lee C, Thomas BC, Ko C, Goff SA, Freeling M (2003) Conserved noncoding sequences in the grasses. Genome Res 13:2030–2041
Katzman S, Kern AD, Bejerano G, Fewell G, Fulton L, Wilson RK, Salama SR, Haussler D (2007) Human genome ultraconserved elements are ultraselected. Science 317:915
Keightley PD, Kryukov GV, Sunyaev S, Halligan DL, Gaffney DJ (2005) Evolutionary constraints in conserved nongenic sequences of mammals. Genome Res 15:1373–1378
Kleinjan DA, van Heyningen V (2005) Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 76:8–32
Kryukov GV, Schmidt S, Sunyaev S (2005) Small fitness effect of mutations in highly conserved non-coding regions. Hum Mol Genet 14:2221–2229
Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE (2007) Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446:926–929
Lettice LA, Horikoshi T, Heaney SJ, van Baren MJ, van der Linde HC, Breedveld GJ, Joosse M, Akarsu N, Oostra BA, Endo N, Shibata M, Suzuki M, Takahashi E, Shinka T, Nakahori Y, Ayusawa D, Nakabayashi K, Scherer SW, Heutink P, Hill RE, Noji S (2002) Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci U S A 99:7548–7553
Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E (2003) A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12:1725–1735
Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA (2000) Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288:136–140
Loots GG, Kneissel M, Keller H, Baptist M, Chang J, Collette NM, Ovcharenko D, Plajzer-Frick I, Rubin EM (2005) Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res 15:928–935
Margulies EH, Blanchette M, Haussler D, Green ED (2003) Identification and characterization of multi-species conserved sequences. Genome Res 13:2507–2518
Masuya H, Sezutsu H, Sakuraba Y, Sagai T, Hosoya M, Kaneda H, Miura I, Kobayashi K, Sumiyama K, Shimizu A, Nagano J, Yokoyama H, Kaneko S, Sakurai N, Okagaki Y, Noda T, Wakana S, Gondo Y, Shiroishi T (2007) A series of ENU-induced single-base substitutions in a long-range cis-element altering Sonic hedgehog expression in the developing mouse limb bud. Genomics 89:207–214
Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046–1047
Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr (2007) Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21:708–718
Nobrega MA, Ovcharenko I, Afzal V, Rubin EM (2003) Scanning human gene deserts for long-range enhancers. Science 302:413
Nobrega MA, Zhu Y, Plajzer-Frick I, Afzal V, Rubin EM (2004) Megabase deletions of gene deserts result in viable mice. Nature 431:988–993
Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM (2006) In vivo enhancer analysis of human conserved non-coding sequences. Nature 444:499–502
Persampieri J, Ritter DI, Lees D, Lehoczky J, Li Q, Guo S, Chuang JH (2008) cneViewer: a database of conserved noncoding elements for studies of tissue-specific gene regulation. Bioinformatics 24:2418–2419
Prabhakar S, Poulin F, Shoukry M, Afzal V, Rubin EM, Couronne O, Pennacchio LA (2006) Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res 16:855–863
Sabherwal N, Bangs F, Roth R, Weiss B, Jantz K, Tiecke E, Hinkel GK, Spaich C, Hauffa BP, van der Kamp H, Kapeller J, Tickle C, Rappold G (2007) Long-range conserved non-coding SHOX sequences regulate expression in developing chicken limb and are associated with short stature phenotypes in human patients. Hum Mol Genet 16:210–222
Sagai T, Masuya H, Tamura M, Shimizu K, Yada Y, Wakana S, Gondo Y, Noda T, Shiroishi T (2004) Phylogenetic conservation of a limb-specific, cis-acting regulator of Sonic hedgehog (Shh). Mamm Genome 15:23–34
Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T (2005) Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 132:797–803
Sakuraba Y, Sezutsu H, Takahasi KR, Tsuchihashi K, Ichikawa R, Fujimoto N, Kaneko S, Nakai Y, Uchiyama M, Goda N, Motoi R, Ikeda A, Karashima Y, Inoue M, Kaneda H, Masuya H, Minowa O, Noguchi H, Toyoda A, Sakaki Y, Wakana S, Noda T, Shiroishi T, Gondo Y (2005) Molecular characterization of ENU mouse mutagenesis and archives. Biochem Biophys Res Commun 336:609–616
Sandelin A, Bailey P, Bruce S, Engstrom PG, Klos JM, Wasserman WW, Ericson J, Lenhard B (2004) Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5:99
Shin JT, Priest JR, Ovcharenko I, Ronco A, Moore RK, Burns CG, MacRae CA (2005) Human-zebrafish non-coding conserved elements act in vivo to regulate transcription. Nucleic Acids Res 33:5437–5445
Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15:1034–1050
Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, Margulies EH, Blanchette M, Siepel AC, Thomas PJ, McDowell JC, Maskeri B, Hansen NF, Schwartz MS, Weber RJ, Kent WJ, Karolchik D, Bruen TC, Bevan R, Cutler DJ, Schwartz S, Elnitski L, Idol JR, Prasad AB, Lee-Lin SQ, Maduro VV, Summers TJ, Portnoy ME, Dietrich NL, Akhter N, Ayele K, Benjamin B, Cariaga K, Brinkley CP, Brooks SY, Granite S, Guan X, Gupta J, Haghighi P, Ho SL, Huang MC, Karlins E, Laric PL, Legaspi R, Lim MJ, Maduro QL, Masiello CA, Mastrian SD, McCloskey JC, Pearson R, Stantripop S, Tiongson EE, Tran JT, Tsurgeon C, Vogt JL, Walker MA, Wetherby KD, Wiggins LS, Young AC, Zhang LH, Osoegawa K, Zhu B, Zhao B, Shu CL, De Jong PJ, Lawrence CE, Smit AF, Chakravarti A, Haussler D, Green P, Miller W, Green ED (2003) Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424:788–793
Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, Strausberg RL, Brenner S (2006) Ancient noncoding elements conserved in the human genome. Science 314:1892
Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA (2008) Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet 40:158–160
Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, Walter K, Abnizova I, Gilks W, Edwards YJ, Cooke JE, Elgar G (2005) Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3:e7
Acknowledgments
We thank all of the technical staff of the Population and Quantitative Genomics Team of RIKEN GSC for the mutation screening. We are also grateful for the computational resources of the RIKEN Super Combined Cluster (RSCC). This research was supported in part by Grants-in-Aid for Scientific Research (C) from the Japan Ministry of Education, Culture, Sports, Science, and Technology (to YS).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Materials
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Sakuraba, Y., Kimura, T., Masuya, H. et al. Identification and characterization of new long conserved noncoding sequences in vertebrates. Mamm Genome 19, 703–712 (2008). https://doi.org/10.1007/s00335-008-9152-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-008-9152-7