Abstract
The basis of eukaryotic complexity is an intricate genetic architecture where parallel systems are involved in tuning gene expression, via RNA-DNA, RNA-RNA, RNA-protein, and DNA-protein interactions. In higher organisms, about 97% of the transcriptional output is represented by noncoding RNA (ncRNA) encompassing not only rRNA, tRNA, introns, 5′ and 3′ untranslated regions, transposable elements, and intergenic regions, but also a large, rapidly emerging family named microRNAs. MicroRNAs are short 20-22-nucleotide RNA molecules that have been shown to regulate the expression of other genes in a variety of eukaryotic systems. MicroRNAs are formed from larger transcripts that fold to produce hairpin structures and serve as substrates for the cytoplasmic Dicer, a member of the RNase III enzyme family. A recent analysis of the genomic location of human microRNA genes suggested that 50% of microRNA genes are located in cancer-associated genomic regions or in fragile sites. This review focuses on the possible implications of microRNAs in post-transcriptional gene regulation in mammalian diseases, with particular focus on cancer. We argue that developing mouse models for deleted and/or overexpressed microRNAs will be of invaluable interest to decipher the regulatory networks where microRNAs are involved.
Similar content being viewed by others
What are microRNAs?
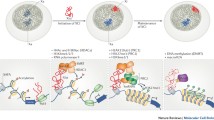
MicroRNAs are a group of small noncoding RNA (ncRNA) molecules, distinct from but related to small interfering RNAs (siRNAs), that have been identified in a variety of organisms (for reviews see He and Hannon 2004; Bartel 2004). These small 20–22-nucleotide (nt) RNAs are transcribed as parts of longer molecules several kilobases (kb) in length that are processed in the nucleus into hairpin RNAs of 70–100 nt by the double-stranded RNA-specific ribonuclease Drosha (Cullen 2004). The hairpin RNAs are transported to the cytoplasm, via an exportin 5-dependent mechanism, where they are digested by a second, double-stranded specific ribonuclease called Dicer. These mechanisms are described in more detail in the next section. In animals, single-stranded microRNA binds specific messenger RNA (mRNA) through sequences that are significantly, though not completely, complementary to the target mRNA, mainly to the 3′ untranslated region (3′ UTR). By a mechanism that is not fully characterized, the bound mRNA remains untranslated, resulting in reduced levels of the corresponding protein; alternatively, the bound mRNA can be degraded, resulting in reduced levels of the corresponding transcript (Fig. 1).
Biogenesis of microRNAs and hypothetical mechanisms in regulation of gene expression. RNA polymerases II and III are believed responsible for microRNA transcription, although a recent publication indicates that polymerase II is the main RNA polymerase for microRNA transcription (Lee et al. 2004). (A) Exonic microRNAs in sense orientation as a part of annotated host genes are transcribed as parts of longer molecules that are processed in the nucleus into hairpin RNAs of 70–100 nt by the dsRNA-specific ribonuclease Drosha. The hairpin RNAs are transported to the cytoplasm where they are digested by a second, double-strand specific ribonuclease called Dicer. In animals, single-stranded miRNA binds specific mRNA through sequences that in most of cases are significantly, though not completely, complementary to the target mRNA. (B) The excision of intronic microRNAs out of the precursors is completed through the process of RNA splicing, followed by Dicer digestion. MicroRNAs are finally incorporated into an RNA-induced silencing complex (RISC) to induce translation suppression or degradation depending of the degree of complementary with the target mRNA. (C, D) Exonic and intronic microRNAs in antisense orientation as a part of annotated host genes can be trancribed as independent transcription units. The mature microRNA sequence, in our hypothetical mechanisms, can lead to translation or transcription suppression of the host gene of other target mRNAs. MicroRNA genes located in intergenic regions or gene deserts are transcribed as independent transcription units and their biogenesis can be described as in A.
The central dogma of classical biology is that genetic information flows from DNA to RNA to proteins. Therefore, “genes” are synonymous with proteins and a gene is defined as a protein-coding region with associated regulatory signals (Mattick 2003). MicroRNAs represent new stars in the gene regulation galaxy, and there is a strong interest among researchers in different fields to understand their mechanism of action and identify their targets. The definition of a gene in the genomics era should be expanded to be a “transcription unit” or “a complete chromosomal segment responsible for making a functional product” (Okazaki et al. 2002). Because of the complex genetic architecture of eukaryotic cells, this definition is still not complete, and additional issues such as overlap, alternative splicing, or transcribed pseudogenes have to be considered (Snyder and Gerstein 2003). Thus, the term “gene” could be expanded to include microRNAs (and other functional ncRNAs).
The number of microRNAs is growing rapidly, especially after using “in silico” cloning (Table 1). One such bioinformatics tool is MiRscan which ranks predicted hairpins that are conserved in the genome of two related animals based on several criteria (Lim et al. 2003). Initially, it was estimated that there could be from 200 to 1000 microRNA genes in the mammalian genome (∼1%–3% of known genes are represented by microRNAs). Today the number of microRNAs, including those electronically cloned, is over 1000 and still growing (Berezikov et al. 2005; Xie et al. 2005). Mammalian microRNA genes have been distinguished by using the prefix mir followed by a number; the prefixes lin and let refer to microRNAs originally identified in C. elegans (see below). The MicroRNA Registry website contains a comprehensive list of microRNAs from all species (Table 1).
How are microRNAs produced?
More than a decade ago, Ambros and colleagues discovered that lin-4, a gene known to control the timing of C. elegans larval development, does not code for any protein but instead codes for a pair of small RNAs (Lee et al. 1993). One RNA is longer (∼70 nt) and can fold in a stem-loop structure, as the precursor of the shorter RNA (∼22 nt). The Ambros and Ruvkun laboratories later discovered that the small lin-4 RNAs had an antisense complementarity to multiple sites in the 3′ UTR of the lin-14 protein-coding gene (Lee et al. 1993; Wightman et al. 1993). The sequence complementarity between the noncoding lin-4 RNA and the 3′ UTR of the protein-coding lin-14 gene suggested that lin-4 regulated translation of lin-14 by binding to the 3′ UTR. It was later shown that lin-4 regulated the lin-14 gene during early larval development as well as the lin-28 gene during late larval development by a mechanism involving RNA-RNA binding that led to suppression of translation (Moss et al. 1997). The shorter lin-4 RNA is now known as the founding member of the class of microRNA genes (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001).
The majority of microRNAs (70%) are located in introns and/or exons, and approximately 30% are located in intergenic regions (Rodriguez et al. 2004) (Fig. 1). The first group of microRNAs from introns and/or exons is oriented in sense with the exon-coding “host” gene and, therefore, may be transcribed as part of annotated genes. The second group of microRNAs are transcribed from intergenic regions or gene deserts (Lagos–Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001), indicating that they form independent transcription units (Lee et al. 2002). The third group of microRNAs are derived from introns and/or exons of annotated genes but are transcribed in the antisense orientation, suggesting that they too form their own transcription units. Tightly linked microRNAs may be transcribed as polycistronic messengers; however, microRNAs separated by more than 50 kb tend to represent independent transcription units (Baskerville and Bartel 2005). RNA polymerases II and III are candidates for pri-microRNAs (primary) transcription (Fig. 1).
In animals (Fig. 1), the first step in microRNA maturation is the nuclear cleavage of the several-kb-long pri-microRNA, which releases an approximately 70-nt, cropped, hairpin-shaped intermediate known as pre-microRNA (precursor). The Drosha RNase III endonuclease is responsible for this nuclear processing (Bartel 2004; Lee et al. 2003; Zeng et al. 2005). Drosha cannot cleave without a partner, forming a complex with DGCR8 (a product of the DiGeorge syndrome critical region gene 8), which contains two double-stranded RNA-binding domains (Han et al. 2004; Tomari and Zamore 2005). The hairpin precursor is actively transported from the nucleus to the cytoplasm by exportin 5 (Yi et al. 2003; Lund et al. 2004). A second enzyme, an RNase III endonuclease called Dicer, is responsible for generating an approximately 21-nt, short, single-stranded RNA that is the mature microRNA (Lee et al. 2003). Dicer was first recognized for its role in generating siRNAs that mediate RNA interference (RNAi) (Bernstein et al. 2001). After cleavage in the cytoplasm, the microRNA pathways of plants and animals share some other steps with RNA silencing, and the cleavage products become incorporated as single-stranded RNAs into the ribonucleoprotein RNA-induced silencing complex RISC (Hammond et al. 2000). The RISC has been purified from fruit fly and human cells and in both cases contains a member of the Argonaute protein family (essential for gene silencing in Caenorhabditis elegans, Neurospora, and Arabidopsis), which is thought to be a core component of the complex (Hammond et al. 2001; Mourelatos et al. 2002). Once incorporated in the cytoplasmic RISC complex, the microRNA will specify cleavage if the mRNA has sufficient complementarity to the microRNA, or it will block translation if the microRNA does not have sufficient complementarity, resulting in reduced expression of the corresponding protein. MicroRNA regulation of the microRNA:mRNA duplex is mediated mainly through multiple complementary sites in the 3′ UTRs, but there are many exceptions. MicroRNAs may also bind the 5′ UTR and/or the coding region of mRNAs, resulting in a similar outcome (Fig. 1).
How are microRNAs working?
The functions of microRNA are various, such as the control of leaf and flower development in plants (Aukerman and Sakai 2003) or the modulation of hematopoietic lineage differentiation in mammals (Chen et al. 2004). Several groups have uncovered roles for microRNAs in the coordination of cell proliferation and cell death during development and in stress resistance and fat metabolism (Ambros 2003). For example, the Drosophila microRNA gene (mir-14) suppresses cell death and is required for normal fat metabolism (Xu et al. 2003), while the bantam locus encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila (Brennecke et al. 2003). It was recently shown that, in addition to their regulatory functions, cellular microRNAs mediate antiviral defenses in human cells (Lecellier et al. 2005).
However, the function of most microRNAs is not known. The antisense single-stranded microRNAs can bind specific mRNA transcripts through sequences that are significantly, though not completely, complementary to the target mRNA. This process is also known as post-transcriptional gene regulation (PTGS). Some microRNAs can downregulate large numbers of target mRNAs (Lim et al. 2005), and it has been speculated that microRNAs could regulate approximately 30% of the human genome (Bartel 2004). MicroRNAs seem to be responsible for fine regulation of gene expression, “tuning” the cellular phenotype during delicate processes like development and differentiation in all organisms, from plants to mammals. Many microRNAs are conserved in sequence between distantly related organisms (see The MicroRNA Registry in Table 1), suggesting that these molecules participate in essential processes (Lagos-Quintana et al. 2003; Pasquinelli et al. 2000). One example is the cluster of mir-16-1 and mir-15a, which is highly conserved in nine of ten primate species (Berezikov et al. 2005).
Target identification has been hampered by the fact that in animals, in contrast to plants, microRNAs do not bind perfectly to their targets. A few nucleotides typically remain unbound, yielding complex secondary structures. Mammalian genes can have more than one microRNA target site in their 3′ UTRs and one microRNA can target more than one mRNA. Bioinformatics approaches have been developed to search for the most thermodynamically favorable microRNA:mRNA duplex interactions (Table 1). Several computational procedures such as DianaMicroT, TargetScan, and miRanda (Lewis et al. 2003; Kiriakidou et al. 2004; Enright et al. 2003) are available to predict microRNA targets. Another set of programs developed for target identification includes RNAhybrid, RNAcalibrate, and RNAeffective (Rehmsmeier et al. 2004), which work by searching for the most energetically favorable hybridization sites for the smaller microRNA within the larger mRNA (Table 1). Only a few target mRNAs have been experimentally proven and studied in vitro, but the numbers of such confirmed interactions are expected to sharply increase as prediction tools become more sophisticated. Generally, the proven mRNA targets are produced from different chromosomes as the corresponding microRNAs (such as mir-19a on chromosome 13q31.3 having as a target the mRNA of the PTEN tumor suppressor gene located on chromosome 10q23.3), but examples of microRNAs targeting “neighbor” exon-coding genes are also known. An important example is that of mir-196, which leads to the cleavage and degradation of mRNA from the homeobox gene HOXB8 (Yekta et al. 2004).
MicroRNAs are not the only ncRNA sequences that have been shown to play a role in the regulation of gene expression. A large set of ncRNAs are known as “gene regulators” and the list in mammals includes Air, H19, Ipw, NTT, Tsix and XIST (Table 2). These ncRNAs have a variety of functions, from potential involvement in the imprinting process to X-chromosome inactivation in mammals. It is likely that other ncRNAs will be found, albeit with additional effects on critical biological processes.
MicroRNAs and human cancer
Several articles have now shown the possibility that small ncRNAs play an important role in cancer (Table 3). We show in Fig. 2 diagrams of several proposed mechanisms for microRNAs as cancer players (Calin et al. 2004a; McManus 2003). Inherited or somatic mutations, amplification, deletion, or epigenetic silencing of microRNA genes may not only cause certain cancers but may contribute to an individual’s risk of developing cancer (Fig. 2).
MicroRNAs as cancer players. MicroRNAs may act as tumor suppressors or oncogenes in cancer. Orange triangles represent mir promoters and blue ovals represent their corresponding mir genes. Orange rectangles represent promoters of protein-coding genes and blue rectangles represent the actual coding sequences of their corresponding genes. One mechanism for the downregulation of “suppressor-microRNAs” that has been identified is (A) Homozygous deletion of microRNA coding regions, as exemplified by deletion of the mir-15a/mir-16-1 cluster in B-CLL (Calin et al. 2002). Hypothetical mechanisms for downregulation of “suppressor-microRNAs” in cancer include (B) the combination of deletion plus promoter hypermethylation, and (C) deletion plus mutation. Mechanisms for the upregulation of “onco-microRNAs” that have been identified are (D) amplification and overexpression of pre-microRNAs, as exemplified by mir-155/BIC in children’s Burkitt’s lymphoma (Metzler et al. 2004), and (E) translocations of either protoconcogenes near the promoter of microRNAs or translocation of microRNAs near the promoters of oncogenes (modified after Calin et al. (©) 2004 PNAS, National Academy of Sciences, Washington, DC, USA).
The first report linking microRNAs and cancer (Calin et al. 2002) involved chronic lymphocytic leukemia (CLL), the most common form of adult leukemia in the Western world. Hemizygous and/or homozygous loss at chromosome 13q14 occurs in more than half of CLL cases. Loss of chromosome 13q14 is also found in more than 50% of mantle cell lymphomas (Stilgenbauer et al. 1998), in approximately 30% of multiple myeloma (Elnenaei et al. 2003), and in approximately 70% of prostate cancers (Dong et al. 2001), suggesting that one or more tumor suppressor genes located at chromosome 13q14 are involved in the pathogenesis of human tumors (Calin et al. 2002). However, detailed genetic analysis, including extensive loss of heterozygosity (LOH), mutation, and expression studies failed to demonstrate the consistent involvement of any of the 12 protein-coding genes located in or close to the deleted region. A cluster of two microRNAs, mir-15a and mir-16-1, was found within the minimal region of deletion (∼30 kb) at 13q14 and to be deleted or downregulated in approximately 70% of CLL samples. Similarly, a t(12;13) chromosome translocation in a patient with CLL was found to decrease the mir-15a and mir-16-1 precursors (Calin et al. 2002). A similar cluster of microRNAs, named mir-15b and mir-16-2, is located on human Chromosome 3 but is expressed at very low levels in lymphoid cells.
The first genome-wide systematic search for correlations between the genomic positions of microRNAs and cancer-associated genomic regions (CAGRs) provided a catalog of microRNAs possibly involved in cancer (Calin et al. 2004a). One hundred eighty-six microRNAs were mapped to the human genome and compared with the location of previously reported nonrandom genetic alterations identified in human tumors and with cloned fragile sites (FRAs). Minimal regions of LOH are suggestive of the presence of tumor suppressor genes, minimal regions of amplification for the presence of oncogenes, while common breakpoint regions are found close to both types of cancer genes. Some common fragile sites predispose to DNA instability in cancer cells; indeed they are preferential sites of sister chromatid exchange, translocation, deletion, amplification, or integration of plasmid DNA and tumor-associated viruses. This study concluded that 19% (35 of 186) of microRNAs are located inside or near FRAs and the relative incidence of microRNAs inside FRAs occurred at a rate nine times higher than in non-FRAs. A significant proportion, 52.5% (98 of 186), of microRNAs are in CAGRs described in a variety of tumors such as lung, breast, ovarian, colon, gastric, and hepatocellular carcinoma, as well as leukemias and lymphomas.
Little was known about the expression levels of microRNA genes in normal and neoplastic cells until 2002. To assess cancer-specific expression levels for hundreds of microRNAs is time-consuming and requires a large amount of total RNA and the use of radioactive isotopes. cDNA microarrays are useful tools for identifying different patterns of expression in a large number of samples. The first developed oligonucleotide microRNA microarray chips (Liu et al. 2004a), containing hundreds of human precursor and mature microRNA probes, identified distinct patterns of microRNA expression in human and mouse tissues (tissue-specific microRNA expression signatures). Another method to determine microRNA expression levels involves the use of a bead-based flow cytometric technique (Lu et al. 2005a). Since the development of these two methods of assessing global microRNA expression, several commercially available platforms have been developed for microRNA gene expression profiling.
Global analysis of microRNA gene expression has shown that these profiles can be used to classify the developmental lineage and differentiation state of tumors; in addition, microRNA profiles appear to provide a distinct signature for tumors which is more precise than that provided by protein-coding genes (Lu et al. 2005a). These findings lead to a unique opportunity to use these microRNA profiles for tumor diagnostics.
Specific differences in microRNA expression between CLL samples and normal CD5+ B cells, the latter representing normal cells corresponding to CLL malignant cells, were found (Calin et al. 2002). Two clusters of CLL samples were identified that were associated with the presence or absence of ZAP-70 expression, a predictor of early disease progression. Low levels of ZAP-70 (identified by both Western blots and flow cytometry) are associated with good prognosis. Two microRNA signatures were associated with the presence or absence of mutations in the expressed immunoglobulin variable-region genes or with deletions at chromosome 13q14, respectively. Different microRNA expression profiles suggested that specific microRNAs can have prognostic significance in CLL, thus expanding the spectrum of prognostic markers (Calin et al. 2004b). A unique microRNA expression signature composed of 13 genes can differentiate cases with low or high ZAP-70 expression and cases with unmutated or mutated IgVH. Furthermore, both mir-15a and mir-16-1 are members of this signature, suggesting an important functional role in the pathogenesis of human CLL (Calin et al. 2005).
The list of microRNAs reported to be involved in cancer is growing continuously (Table 3). One report showed that mir-143 and mir-145 exhibited reduced levels of the mature microRNA in colon cancer patients, colon cancer cell lines, and precancerous adenomatous polyps when compared to normal tissues by Northern blot (Michael et al. 2003). On the contrary, levels of unprocessed hairpin precursors were not altered. It is possible that the reduced accumulation of the mature microRNA reflects early changes in neoplastic cells or that this depletion is related to their activity in binding and inactivating target mRNAs in the cytoplasm. Putative targets (human mRNAs that display imperfect complementarity of 70%–100%) for mir-143 and mir-145 include several genes that encode components of signal transduction pathways (such as RAF, RHO, and G-proteins) and chromatin-mediated control of gene expression were found by computer search (Table 3).
In an independent study, it was shown that mir-143 regulates adipocyte differentiation (Esau et al. 2004). When antisense oligonucleotides (ASOs) targeting human microRNAs were transfected into cultured human primary subcutaneous preadipocytes, mir-143 was upregulated as adipocytes differentiated. Bioinformatics approaches predicted several putative microRNA targets. ERK5/BMK1 (which promotes cell growth and proliferation in response to tyrosine kinase signaling) was predicted to be a mir-143 target and, in fact, ERK5 protein levels were upregulated in mir-143 ASO-treated adipocytes. However, because each microRNA can regulate multiple target genes, mir-143 could directly inhibit ERK5 by binding the 3′ UTR of ERK5, or indirectly by binding to another target gene (Esau et al. 2004).
Another study reported reduced expression of the let-7 microRNA in human lung cancer and the correlation with clinicopathologic features (Takamizawa et al. 2004). Reduction in the expression (>80%) levels of let-7 was observed in 60% (12 of 20) of cancer cell lines and 44% (7 of 16) of lung tumors when compared with normal lung tissues by Northern blots. The reduced expression was significantly associated with shortened survival of patients after potentially curative resection. One hundred forty-three cases had been followed for more than five years after surgery and were used to study the prognostic significance of let-7. One hundred forty-three non-small-cell lung carcinoma (NSCLC) cases were classified into clusters 1 and 2 (cluster 1 with low let-7 expression and higher disease stages; cluster 2 with high let-7 expression and lower disease stages). The difference in postoperative survival between clusters 1 and 2 was highly significant. In addition, overexpression of let-7 in the A549 lung adenocarcinoma cell line inhibited lung cancer cell growth in vitro (Takamizawa et al. 2004). Several observations led to the conclusion that let-7 acts as a tumor suppressor gene in lung tissue. Several let-7 family members, including let-7a-2, let-7c, and let-7g, have been mapped to minimally deleted regions in lung cancers (Calin et al. 2004a). Let-7 expression is lower in lung tumors than in normal lung tissue, while RAS protein is significantly higher in lung tumors (Johnson et al. 2005). Finally, expression of the RAS oncogene is regulated by let-7, and overexpression of let-7 has been shown to inhibit lung tumor cell line growth (Johnson et al. 2005).
High expression of human BIC RNA has been found in Hodgkin’s lymphoma (van den Berg 2003). In contrast, an analysis of non-Hodgkin’s lymphoma (NHL) did not reveal any remarkable upregulation of BIC expression. The BIC locus was originally identified as a common retroviral integration site in avian leukosis virus-induced B-cell lymphomas (Tam et al. 1997). The human BIC locus also encodes a microRNA, mir-155 (Tam 2001; Lagos-Quintana et al. 2002). The conditio sine qua non for the development of Burkitt’s lymphoma is activation of the MYC oncogene, mostly by chromosomal translocations in which MYC is juxtaposed next to an immunoglobulin enhancer. But activation of MYC alone is not sufficient for full malignancy; in fact, MYC cooperates with other oncogenes. Further studies involving a human B-cell line that contains a tetracycline-regulated c-MYC gene have shown that overexpression of the c-MYC oncogene leads to increased expression of six microRNAs, two of which are mir-17-5p and mir-20a (O’Donnell et al. 2005). These microRNAs were found to negatively regulate expression of the E2F1 transcription factor. Since E2F1 is also a target of c-MYC, these results allude to an intricate network that exquisitely regulates gene expression, ultimately controlling cell proliferation.
In childhood Burkitt’s lymphoma, there has been reported a 100-fold upregulation of the hairpin precursor mir-155 (Metzler et al. 2004). It could be speculated that mir-155 directly downregulates one of the MYC antagonists such as MAD1, MXI1, or ROX/MNT. Accumulation of mir-155 and BIC RNA was also reported in other B-cell lymphomas (Eis et al. 2005).
It was recently reported that mir-21 is upregulated in both human glioblastomas as well as cultured glioblastoma multiforma cells and cell lines compared to primary astrocyte cultures derived from normal tissue; inhibition of mir-21 expression led to the activation of caspases and cell death in vitro (Chan et al. 2005). These data suggest a role for mir-21 in malignant glioblastoma, most likely by influencing expression of genes involved in apoptosis (Chan et al. 2005).
Overall, the studies to date clearly implicate a causative role for microRNAs in human cancer. The intricate network of signals regulating gene expression requires a delicate balance between protein-coding genes and microRNA genes. Disruption of this delicate relationship by mutation can tip the balance in favor of the development of cancer.
MicroRNAs and other human diseases
Several articles have been published showing a probable link between microRNAs and other human diseases, but the precise mechanisms are still not known (Table 4). Fragile X syndrome is the most common inherited mental retardation disease. In the U.S., conservative estimates report that fragile X syndrome affects approximately 1 in 4000 males and 1 in 8000 females; the rate of the female carrier state has been estimated to be as high as 1 in 250 and the male carrier state has been estimated to be 1 in 1000. The fragile X mental retardation protein (FMRP) is an RNA-binding protein that can function as a translational suppressor. FMRP can form a messenger ribonucleoprotein (mRNP) complex, interacting with specific RNA transcripts and proteins. The complex is transported out of the nucleus and into the cytoplasm, where it can either associate with the ribosome or interact with RISC; in both cases, it will lead to protein synthesis regulation. FMRP can interact with microRNAs and other components of the microRNA pathways, including Dicer and the mammalian ortholog of Argonaute 1 (AGO1), a component of RISC (Jin et al. 2004; Peng et al. 2004).
Spinal muscular atrophy (SMA) is an inherited neurodegenerative disease with neuromuscular symptoms. Survival of Motor Neurons protein (SMN) is a component of the SMN complex that has critical functions in the assembly and restructuring of diverse ribonucleoprotein (RNP) complexes. Components of the SMN complex (GEMIN3 and GEMIN4) are also in a separate complex that contains eIF2C2, a member of the Argonaute protein family. This novel complex contains numerous microRNAs that form novel RNPs named miRNPs (Mourelatos et al. 2002). This link between microRNAs and a devastating neurodegenerative disease is intriguing and will be of great interest to determine what effect these microRNAs have on miRNPs in SMA patients.
DiGeorge syndrome is characterized by a few specific cardiac malformations, facial deformity, and certain endocrine and immune anomalies. The DGCR8 (DiGeorge syndrome critical region gene 8) protein and Drosha interact in human cells. Knockdown of the Drosophila dmDGCR8 resulted in a 5–23-fold accumulation of some pri-microRNAs, confirming a role in mediating the genesis of miRNAs from the pri-microRNA transcript (Landthaler et al. 2004; Gregory et al. 2004).
MicroRNAs have also been shown to regulate insulin secretion (Poy et al. 2004). Overexpression of mir-375 (a pancreatic islet-specific microRNA) suppresses glucose-induced insulin secretion, and, conversely, inhibition of endogenous mir-375 by ASOs enhances insulin secretion in murine pancreatic α- and β-cell lines. Bioinformatics approaches predicted several putative targets with a potential role in insulin secretion such as MAPK4, MKI1, VTI1A, and MTPN; the last two proteins are known to be involved in vesicle transport of neurons and in neurotransmitter release.
These studies represent the beginning links between microRNAs and human disease. Further investigations are likely to reveal the involvement of additional microRNAs and their targets in simple and complex genetic diseases.
Mouse models for microRNA studies
Almost all mammalian microRNAs are highly conserved and more than 220 mouse microRNAs were cloned by conventional methods (Lagos–Quintana et al. 2002; Seitz et al. 2004) (see The MicroRNA Registry in Table 1). It has been shown that a tissue-specific microRNA dominates the population of expressed microRNAs, suggesting important roles in tissue differentiation. For example, in mouse liver, mir-122a and mir-122b represent 72% of all cloned microRNAs; these microRNAs were not detected in the other tissues analyzed. In spleen, mir-143 was the most abundant microRNA. In colon, mir-142 was cloned several times; however, because of the strong RNase activity in small intestine and pancreas, too few microRNA sequences were obtained from these tissues to reach statistical significance.
Using a microRNA microarray approach, microRNA expression was analyzed in 17 mouse organs and tissues (Babak et al. 2004). More than half of the 78 microRNAs detected were expressed in specific adult tissues. The results were confirmed by Northern blotting and were consistent with microRNA expression data reported in the literature. Furthermore, this study is consistent with microRNA expression profiling in human tissues (Liu et al. 2004). For example, seven of eight microRNAs were brain-specific in both mice and humans.
Another report gave new light on the world of microRNA regulation in humans and mice (Seitz et al. 2004). By bioinformatics approaches, 46 potential microRNAs located in the human imprinted 14q32 domain have been identified. Forty of these microRNAs are clustered and most of them are arranged in tandem arrays. The majority of these genes map within an approximately 40-kb region, making it the largest microRNA cluster that has been described. In mouse, this microRNA cluster is conserved at the homologous region on Chromosome 12. The detected microRNAs are expressed in embryos and placenta, whereas in the adult their expression pattern is limited to the brain. Expression of these microRNAs comes solely from the maternally inherited chromosome, which is imprinted. Regulation of their expression is dictated by a region located approximately 200 kb upstream from the microRNA cluster (Seitz et al. 2004).
Research has shown how microRNAs have a role in fine-tuning specific Hox mRNA expression patterns during mouse development (Mansfield et al. 2004). A novel approach was used to monitor microRNA tissue-specific expression patterns during embryogenesis. Until now, microRNA expression studies were done by Northern blots, tissue-specific RNA cloning, and microarrays, but all of these methods are not useful for displaying spatiotemporal expression patterns. In situ hybridization methods are still difficult to adapt in vertebrate embryos because of the small size of microRNAs. An alternative approach, previously used in Drosophila melanogaster, was used where several reporter transgenes (“sensors”) were made to detect microRNAs in the embryo. The sensor was a constitutively expressed reporter gene (lacZ) containing microRNA complementary sequences in the 3′ UTR. In cells lacking the corresponding microRNAs, the reporter transgene is stable and will express b-galactosidase. In contrast, cells expressing the corresponding microRNA degrade the lacZ message by an RNA interfering pathway (RNAi), thus resulting in absence of b-galactosidase activity.
To prove the biological significance of microRNAs in human biology and in cancer in particular, the development of mouse models for deleted and/or overexpressed microRNAs will be of invaluable interest for deciphering the regulatory networks where microRNAs are involved. Several groups have started this task by disrupting Dicer1 in mice. Loss of Dicer1 by replacement of exon 21 with a neomycin-resistance cassette leads to lethality early in embryogenesis at day 7.5 of development; Dicer1-null embryos were found to be depleted of pluripotent stem cells (Bernstein et al. 2003). Another group generated Dicerex1/2 mice that have a deletion of the amino acid sequences from the first and second exons of the dicer gene (Yang et al. 2005). Dicerex1/2 homozygous embryos died between days 12.5 and 14.5 of gestation, again demonstrating that Dicer is necessary for normal mouse development. In addition, blood vessel formation and maintenance in Dicerex1/2 embryos and yolk sacs were severely compromised, with altered expression of Vegf, Flt1, Kdr, and Tie1 being detected in the mutant embryos. This study suggests that Dicer has a role in embryonic angiogenesis, probably through processing of microRNAs that regulate expression levels of key angiogenic regulators (Yang et al. 2005).
Although Dicer-null embryonic stem (ES) cells are viable, they are defective in RNA interference (RNAi) and the generation of microRNAs (Kanellopoulou et al. 2005). These mutant ES cells displayed severe defects in differentiation both in vitro and in vivo. An increase in centromeric repeat transcripts was detected, whereas the expression of homologous small double-stranded RNAs was markedly reduced in Dicer-deficient ES cells (Kanellopoulou et al. 2005; Murchison et al. 2005). Re-expression of Dicer in the knockout cells rescued these phenotypes. These Dicer-deficient cells were able to undergo gene silencing when provided with synthetic siRNAs (Kanellopoulou et al. 2005; Murchison et al. 2005), indicating that Dicer function is required for processing of microRNAs but not required for the action of microRNAs once they are produced. However, the status of cytosine methylation of centromeric satellite sequences remains in question. Taken together, these studies suggest that the essential role of Dicer in the production of mature microRNAs is central for biological processes in mammalian organisms, ranging from stem cell differentiation to the integrity and function of centromeric heterochromatin (Kanellopoulou et al. 2005).
Critical insights into the roles of individual proteins of the microRNA processing machinery have been provided by in situ hybridization studies of Argonaute (Ago1-Ago4) family members in mouse embryos (Liu et al. 2004b; Lu et al. 2005b). Studies in embryos at days 9.5–14.5 of development have shown that Argonaute family members are restricted in both their temporal and spatial patterns of expression. However, Ago2 shows the broadest expression in mouse embryos and is consistent with the fact that Ago2-deficient mice die early in development (Liu et al. 2004b). Thus, the differential expression of Ago family members suggests specific roles for these proteins during organogenesis, most likely involving the production of microRNAs that are controlled at different times and in specific tissues by the presence of Ago proteins.
One example of the use of mouse models to study the roles of specific microRNAs in cancer was provided by He et al. (2005). Their initial observation was that the human mir 17-92 cluster was overexpressed by more than 5-fold in a majority of human B-cell lymphomas. To determine whether overexpression of this cluster could drive B-cell lymphomagenesis in mouse, the authors derived hematopoeitic stem cells from a susceptible Eμ-myc transgenic mouse and transduced them with a retrovirus carrying the mir 17-92 cluster. Injection of transduced cells into normal mice resulted in acceleration of the development of B-cell lymphomas; in contrast, other subsets of microRNAs expressed in these cells did not have similar effects (He et al. 2005).
The ability to manipulate the genome of mice coupled with the multitude of inbred strain backgrounds provides invaluable tools to genetically dissect the biological functions of individual microRNAs. Further studies of mouse models will provide significant insights into the roles of different microRNAs in development and disease processes.
Conclusions
A link between microRNAs and human diseases, in particular, with cancer as focused on in this review, has been recently shown. To better understand the biological significance of microRNAs and of alterations found in human diseases (such as the effect of microRNA mutations or variations in expression) the development of mouse models is mandatory. Regulation of gene expression in the mammalian genome, during development, differentiation, and disease, is a complex and multitasked system. To make transcripts instead of proteins is energetically “less expensive” for cells and could be the reason why regulation at the RNA level is “cheaper” and more efficient than at the protein level. Penetrance in gene expression requires a fine regulation and microRNAs could have a role in different phenotypic expressions of the same gene. The ability to simultaneously regulate large sets of genes by a single microRNA appears to be at the heart of control of multiple pathways that include morphogenesis and cell fate decisions, response to infectious organisms, and centromeric heterochromatin structure. MicroRNAs appear to be at the center of the balance between apoptosis and proliferation in cancer cells and thus could provide novel approaches for therapeutics. MicroRNAs are small stars in the genome galaxy with many surprises still in store.
References
Ambros V (2003) MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113: 673–676
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741
Avner P, Heard E (2001) X-chromosome inactivation: counting, choice and initiation. Nat Rev Genet 2: 59–67
Babak T, Zhang W, Morris Q, Blencowe BJ, Hughes TR (2004) Probing microRNAs with microarrays: tissue specificity and functional inference. RNA 10: 1813–1819
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 23: 281–297
Baskerville S, Bartel DP (2005) Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11, 241–247
Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, et al. (2005) Phylogenetic shadowing and computational identification of human microRNA genes. Cell 14: 21–24
Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366
Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, et al. (2003) Dicer is essential for mouse development. Nat Genet 35: 215–217
Brannan CI, Dees EC, Ingram RS, Tilghman SM (1990) The product of the H19 gene may function as an RNA. Mol Cell Biol 10: 28–36
Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, et al. (2002) Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 99: 15524–15529
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, et al. (2004a) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 101: 2999–3004
Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, et al. (2004b) MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci U S A 101: 11755–11760
Calin GA, Ferracin M, Cimino A, Di Leva G, Shimizu M, et al. (2005) A unique microRNA signature associated with prognostic factors and disease progression in B cell chronic lymphocytic leukemia. N Engl J Med 353: 1793–1801
Chan JA, Krichevsky AM, Kosik KS (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65: 6029–6033
Chen CZ, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303: 83–86
Cullen BR (2004) Transcription and processing of human microRNA precursors. Mol Cell 16: 861–865
Dong JT, Boyd JC, Frierson HF Jr (2001) Loss of heterozygosity at 13q14 and 13q21 in high grade, high stage prostate cancer. Prostate 49: 166–171
Eis PS, Tam W, Sun L, Chadburn A, Li Z, et al. (2005) Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A 102: 3627–3632
Elnenaei MO, Hamoudi RA, Swansbury J, Gruszka-Westwood AM, Brito-Babapulle V, et al. (2003) Delineation of the minimal region of loss at 13q14 in multiple myeloma. Genes Chromosomes Cancer 36: 99–106
Enright AJ, John B, Gaul U, Tuschl T, Sander C, et al. (2003) MicroRNA targets in Drosophila. Genome Biol 5: RI.1–14
Erdmann VA, Szymanski M, Hochberg A, de Groot N, Barciszewski J (2000) Non-coding, mRNA-like RNAs database Y2K. Nucleic Acids Res 28: 197–200
Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, et al. (2004) MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 50: 52361–52365
Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, et al. (2004) The microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240
Hammond S, Bernstein E, Beach D, Hannon G (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–229
Hammond SM, Caudy AA, Hannon GJ (2001) Post-transcriptional gene silencing by double-stranded RNA. Nature Rev Gen 2: 110–119
Han J, Lee Y, Yeom KH, Kim YK, Jin H (2004) The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, et al. (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833
Jin P, Alisch RS, Warren ST (2004) RNA and microRNAs in fragile X mental retardation. Nat Cell Biol 6: 1048–1053
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, et al. (2005) RAS is regulated by the let-7 microRNA family. Cell 120: 635–647
Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, et al. (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501
Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, et al. (2004) A combined computational-experimental approach predicts human microRNA targets. Genes Dev 18: 1165–1178
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294: 853–858
Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, et al. (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735–739
Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T (2003) New microRNAs from mouse and human. RNA 9: 175–179
Landthaler M, Yalcin A, Tuschl T (2004) The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862
Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, et al. (2005) A cellular microRNA mediates antiviral defense in human cells. Science 308: 557–560
Lee RC, Ambrose V (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854
Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J 21: 4663–4670
Lee Y, Ahn C, Han J, Choi H, Kim J (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419
Lee Y, Kim M, Han J, Yeom KH, Lee S, et al. (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115: 787–98
Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP (2003) Vertebrate microRNA genes. Science 299: 1540
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, et al. (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433: 769–773
Liu AY, Torchia BS, Migeon BR, Siliciano RF (1997) The human NTT gene: identification of a novel 17-kb noncoding nuclear RNA expressed in activated CD4+ T cells. Genomics 39: 171–184
Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, et al. (2004a) An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A 101: 9740–9744
Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, et al. (2004b) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441
Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, et al. (2002) Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 23: 1885–1895
Lu J, Getz G, Miska EA, Alvarez–Saavedra E, Lamb J, et al. (2005a) MicroRNA expression profiles classify human tumors. Nature 435: 834–838
Lu J, Qian J, Chen F, Tang XZ, Li C, et al. (2005b) Differential expression of components of the microRNA machinery during mouse organogenesis. Biochem Biophys Res Commun 334: 319–323
Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U (2003) Nuclear export of microRNA precursors. Science 303: 95–98
Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, et al. (2004) MicroRNA-responsive “sensor” transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet 36: 1079–1083
Mattick JS (2003) Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 25: 930–939
McManus MT (2003) MicroRNAs and cancer. Semin Cancer Biol 13: 253–258
Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A (2004) High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer 39: 167–169
Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ (2003) Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 1: 882–891
Moss EG, Lee RC, Ambros V (1997) The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88: 637–646
Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, et al. (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev 16: 720–728
Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ (2005) Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A 34: 12135–12140
O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-MYC-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843
Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bano H, et al. (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420, 563–573
Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, et al. (2004) Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res 64: 3087–3095
Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, et al. (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408: 86–89
Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, et al. (2004) A pancreatic islet-specific microRNA regulates insulin secretion. Nature 432: 226–230
Ratter BP, Meller VH (2004) Drosophila male-specific lethal 2 protein controls sex-specific expression of the roX genes. Genetics 166: 1825–1832
Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10: 1507–1517
Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14: 1902–1910
Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC (2004) A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res 14: 1741–1748
Sleutels F, Zwart R, Barlow DP (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813
Snyder M, Gerstein M (2003) Genomics. Defining genes in the genomics era. Science 300: 258–260
Stilgenbauer S, Nickolenko J, Wilhelm J, Wolf S, Weitz S, et al. (1998) Expressed sequences as candidates for a novel tumor suppressor gene at band 13q14 in B-cell chronic lymphocytic leukemia and mantle cell lymphoma. Oncogene 16: 1891–1897
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H (2004) Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64: 3753–3756
Tam W (2001) Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene 274: 157–167
Tam W, Ben-Yehuda D, Hayward WS (1997) BIC, a novel gene activated by proviral insertions in avain leucosis virus-induced lymphomas, is likely to function through its non coding RNA. Mol Cell Biol 17: 1490–1502
Tomari Y, Zamore PD (2005) MicroRNA biogenesis: Drosha can’t cut it without a partner. Curr Biol 15: R61–R64
van den Berg A, Kroesen BJ, Kooistra K, de Jong D, Briggs J, et al. (2003) High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer 37: 20–28
Wevrick R, Francke U (1997) An imprinted mouse transcript homologous to the human imprinted in Prader-Willi syndrome (IPW) gene. Hum Mol Genet 6: 325–332
Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862
Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, et al. (2005) Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345
Xu P, Vernooy SY, Guo M, Hay BA (2003) The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol 13: 790–795
Yang T, Adamson TE, Resnick JL, Leff S, Wevrick R, et al. (1998) A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet 19: 25–31
Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, et al. (2005) Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem 280: 9330–9335
Yekta S, Shih I-H, Bartel DP (2004) MicroRNA-directed cleavage of HOXB8 mRNA. Science 304: 594–595
Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016
Zeng Y, Yi R, Cullen BR (2005) Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J 24: 138–148
Acknowledgments
The authors thank Dr. Marina Markova for reviewing the manuscript. This work was supported by Program Project Grants P01CA76259, P01CA81534, and P30CA56036 from the National Cancer Institute to CMC. GAC was the recipient of a Kimmel Scholar award. LDS was supported by NCI grants PO1 CA72027 and RO1 CA89560.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Sevignani, C., Calin, G.A., Siracusa, L.D. et al. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome 17, 189–202 (2006). https://doi.org/10.1007/s00335-005-0066-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-005-0066-3