Abstract
Objective
To assess the performance of convolutional neural networks (CNNs) for semiautomated segmentation of hepatocellular carcinoma (HCC) tumors on MRI.
Methods
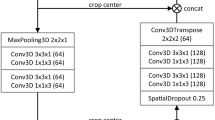
This retrospective single-center study included 292 patients (237 M/55F, mean age 61 years) with pathologically confirmed HCC between 08/2015 and 06/2019 and who underwent MRI before surgery. The dataset was randomly divided into training (n = 195), validation (n = 66), and test sets (n = 31). Volumes of interest (VOIs) were manually placed on index lesions by 3 independent radiologists on different sequences (T2-weighted imaging [WI], T1WI pre-and post-contrast on arterial [AP], portal venous [PVP], delayed [DP, 3 min post-contrast] and hepatobiliary phases [HBP, when using gadoxetate], and diffusion-weighted imaging [DWI]). Manual segmentation was used as ground truth to train and validate a CNN-based pipeline. For semiautomated segmentation of tumors, we selected a random pixel inside the VOI, and the CNN provided two outputs: single slice and volumetric outputs. Segmentation performance and inter-observer agreement were analyzed using the 3D Dice similarity coefficient (DSC).
Results
A total of 261 HCCs were segmented on the training/validation sets, and 31 on the test set. The median lesion size was 3.0 cm (IQR 2.0–5.2 cm). Mean DSC (test set) varied depending on the MRI sequence with a range between 0.442 (ADC) and 0.778 (high b-value DWI) for single-slice segmentation; and between 0.305 (ADC) and 0.667 (T1WI pre) for volumetric-segmentation. Comparison between the two models showed better performance in single-slice segmentation, with statistical significance on T2WI, T1WI-PVP, DWI, and ADC. Inter-observer reproducibility of segmentation analysis showed a mean DSC of 0.71 in lesions between 1 and 2 cm, 0.85 in lesions between 2 and 5 cm, and 0.82 in lesions > 5 cm.
Conclusion
CNN models have fair to good performance for semiautomated HCC segmentation, depending on the sequence and tumor size, with better performance for the single-slice approach. Refinement of volumetric approaches is needed in future studies.
Key Points
• Semiautomated single-slice and volumetric segmentation using convolutional neural networks (CNNs) models provided fair to good performance for hepatocellular carcinoma segmentation on MRI.
• CNN models’ performance for HCC segmentation accuracy depends on the MRI sequence and tumor size, with the best results on diffusion-weighted imaging and T1-weighted imaging pre-contrast, and for larger lesions.





Similar content being viewed by others
Abbreviations
- ADC:
-
Apparent diffusion coefficient
- AI:
-
Artificial intelligence
- AP:
-
Arterial phase
- CNN:
-
Convolutional neural network
- CT:
-
Computed tomography
- DL:
-
Deep learning
- DP:
-
Delayed phase
- DSC:
-
Dice similarity coefficient
- DWI:
-
Diffusion-weighted imaging
- HBP:
-
Hepatobiliary phase
- HCC:
-
Hepatocellular carcinoma
- LI-RADS:
-
Liver Imaging Reporting and Data System
- MRI:
-
Magnetic resonance imaging
- PVP:
-
Portal venous phase
- T1WI:
-
T1-weighted imaging
- T2WI:
-
T2-weighted imaging
- VOI:
-
Volume of interest
References
Vietti Violi N, Lewis S, Hectors S, Said D, Taouli B (2019) Radiological diagnosis and characterization of HCC. In: Hoshida Y, (ed) Hepatocellular carcinoma: translational precision medicine approaches, Cham (CH), pp 71–92
Cunha GM, Sirlin CB, Fowler KJ (2021) Imaging diagnosis of hepatocellular carcinoma: LI-RADS. Chin Clin Oncol 10:3
Gordic S, Corcuera-Solano I, Stueck A et al (2017) Evaluation of HCC response to locoregional therapy: validation of MRI-based response criteria versus explant pathology. J Hepatol 67:1213–1221
Llovet JM, Lencioni R (2020) mRECIST for HCC: performance and novel refinements. J Hepatol 72:288–306
Forner A, Ayuso C, Varela M et al (2009) Evaluation of tumor response after locoregional therapies in hepatocellular carcinoma: are response evaluation criteria in solid tumors reliable? Cancer 115:616–623
Suzuki C, Torkzad MR, Jacobsson H et al (2010) Interobserver and intraobserver variability in the response evaluation of cancer therapy according to RECIST and WHO-criteria. Acta Oncol 49:509–514
Sohaib SA, Turner B, Hanson JA, Farquharson M, Oliver RT, Reznek RH (2000) CT assessment of tumour response to treatment: comparison of linear, cross-sectional and volumetric measures of tumour size. Br J Radiol 73:1178–1184
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Nougaret S, Vargas HA, Lakhman Y et al (2016) Intravoxel incoherent motion-derived histogram metrics for assessment of response after combined chemotherapy and radiation therapy in rectal cancer: initial experience and comparison between single-section and volumetric analyses. Radiology 280:446–454
van Heeswijk MM, Lambregts DM, van Griethuysen JJ et al (2016) Automated and semiautomated segmentation of rectal tumor volumes on diffusion-weighted MRI: can it replace manual volumetry? Int J Radiat Oncol Biol Phys 94:824–831
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444
Becker AS, Marcon M, Ghafoor S, Wurnig MC, Frauenfelder T, Boss A (2017) Deep learning in mammography: diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer. Invest Radiol 52:434–440
Drozdzal M, Chartrand G, Vorontsov E et al (2018) Learning normalized inputs for iterative estimation in medical image segmentation. Med Image Anal 44:1–13
Trebeschi S, van Griethuysen JJM, Lambregts DMJ et al (2017) Deep learning for fully-automated localization and segmentation of rectal cancer on multiparametric MR. Sci Rep 7:5301
Vorontsov E, Cerny M, Régnier P et al (2019) Deep learning for automated segmentation of liver lesions at CT in patients with colorectal cancer liver metastases. Radiol Artif Intell 1:180014
Isensee F, Petersen J, Klein A et al (2018) nnu-net: self-adapting framework for u-net-based medical image segmentation. arXiv preprint arXiv:180910486
Hu P, Wu F, Peng J, Liang P, Kong D (2016) Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol 61:8676–8698
Lu F, Wu F, Hu P, Peng Z, Kong D (2017) Automatic 3D liver location and segmentation via convolutional neural network and graph cut. Int J Comput Assist Radiol Surg 12:171–182
van Gastel MDA, Edwards ME, Torres VE, Erickson BJ, Gansevoort RT, Kline TL (2019) Automatic Measurement of kidney and liver volumes from mr images of patients affected by autosomal dominant polycystic kidney disease. J Am Soc Nephrol 30:1514–1522
Wang K, Mamidipalli A, Retson T et al (2019) Automated CT and MRI liver segmentation and biometry using a generalized convolutional neural network. Radiol Artif Intell 1:180022
Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S (2018) Liver fibrosis: deep convolutional neural network for staging by using gadoxetic acid-enhanced hepatobiliary Phase MR images. Radiology 287:146–155
Choi KJ, Jang JK, Lee SS et al (2018) Development and validation of a deep learning system for staging liver fibrosis by using contrast agent-enhanced CT images in the liver. Radiology 289:688–697
Wang K, Lu X, Zhou H et al (2019) Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut 68:729–741
Bousabarah K, Letzen B, Tefera J et al (2020) Automated detection and delineation of hepatocellular carcinoma on multiphasic contrast-enhanced MRI using deep learning. Abdom Radiol (NY). https://doi.org/10.1007/s00261-020-02604-5
Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286:887–896
Hamm CA, Wang CJ, Savic LJ et al (2019) Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol. https://doi.org/10.1007/s00330-019-06205-9
Chartrand G, Cheng PM, Vorontsov E et al (2017) Deep learning: a primer for radiologists. Radiographics 37:2113–2131
Li X, Morgan PS, Ashburner J, Smith J, Rorden C (2016) The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J Neurosci Methods 264:47–56
Tustison NJ, Avants BB, Cook PA et al (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320
Chernyak V, Fowler KJ, Kamaya A et al (2018) Liver Imaging Reporting and Data System (LI-RADS) version 2018: imaging of hepatocellular carcinoma in at-risk patients. Radiology 289:816–830
Zunair H, Ben Hamza A (2021) Sharp U-Net: depthwise convolutional network for biomedical image segmentation. Comput Biol Med 136:104699
Lowekamp BC, Chen DT, Ibanez L, Blezek D (2013) The Design of SimpleITK. Front Neuroinform 7:45
Besa C, Kakite S, Cooper N, Facciuto M, Taouli B (2015) Comparison of gadoxetic acid and gadopentetate dimeglumine-enhanced MRI for HCC detection: prospective crossover study at 3 T. Acta Radiologica Open 4:2047981614561285
ViettiVioli N, Argiriadi P, Rosen A et al (2020) Gadoxetate disodium-enhanced MRI: Assessment of arterial phase artifacts and hepatobiliary uptake in a large series. Eur J Radiol 132:109313
Brismar TB, Dahlstrom N, Edsborg N, Persson A, Smedby O, Albiin N (2009) Liver vessel enhancement by Gd-BOPTA and Gd-EOB-DTPA: a comparison in healthy volunteers. Acta Radiol 50:709–715
Tamada T, Ito K, Sone T et al (2009) Dynamic contrast-enhanced magnetic resonance imaging of abdominal solid organ and major vessel: comparison of enhancement effect between Gd-EOB-DTPA and Gd-DTPA. J Magn Reson Imaging 29:636–640
Vorontsov E, Cerny M, Regnier P et al (2019) Deep learning for automated segmentation of liver lesions at CT in patients with colorectal cancer liver metastases. Radiol Artif Intell 1:180014
Ouhmich F, Agnus V, Noblet V, Heitz F, Pessaux P (2019) Liver tissue segmentation in multiphase CT scans using cascaded convolutional neural networks. Int J Comput Assist Radiol Surg 14:1275–1284
Lewis S, Hectors S, Taouli B (2021) Radiomics of hepatocellular carcinoma. Abdom Radiol (NY) 46:111–123
Said D, Hectors SJ, Wilck E et al (2020) Characterization of solid renal neoplasms using MRI-based quantitative radiomics features. Abdom Radiol (NY) 45:2840–2850
Funding
This work was partially funded by Owkin Inc.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Bachir Taouli.
Conflict of interest
Daniela Said: None to declare relating to this study.
Guillermo Carbonell: None to declare relating to this study.
Daniel Stocker: None to declare relating to this study.
Naik Vietti Violi: None to declare relating to this study.
Octavia Bane: None to declare relating to this study.
Xing Chin: None to declare relating to this study.
Myron Schwartz: None to declare relating to this study.
Parissa Tabrizian: None to declare relating to this study.
Sara Lewis: None to declare relating to this study.
Daniel Stocker: None to declare relating to this study.
Stefanie Hectors: Employee of Regeneron.
Hayit Greenspan: None to declare relating to this study.
Jean-Baptiste Schiratti: Employee of Owkin.
Simon Jégou: Employee of Owkin.
Paul Jehanno: Employee of Owkin.
Bachir Taouli: Consultancy and/or advisory roles for Bayer, Guerbet, and Helio Health and research funding/support from Bayer, Takeda, Regeneron, Siemens, and Echosens.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Study subjects or cohorts overlap
None.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Said, D., Carbonell, G., Stocker, D. et al. Semiautomated segmentation of hepatocellular carcinoma tumors with MRI using convolutional neural networks. Eur Radiol 33, 6020–6032 (2023). https://doi.org/10.1007/s00330-023-09613-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-023-09613-0