Abstract
Objectives
To assess whether integrative radiomics and transcriptomics analyses could provide novel insights for radiomic features’ molecular annotation and effective risk stratification in non-small cell lung cancer (NSCLC).
Methods
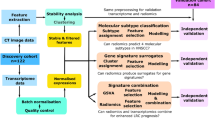
A total of 627 NSCLC patients from three datasets were included. Radiomics features were extracted from segmented 3-dimensional tumour volumes and were z-score normalized for further analysis. In transcriptomics level, 186 pathways and 28 types of immune cells were assessed by using the Gene Set Variation Analysis (GSVA) algorithm. NSCLC patients were categorized into subgroups based on their radiomic features and pathways enrichment scores using consensus clustering. Subgroup-specific radiomics features were used to validate clustering performance and prognostic value. Kaplan–Meier survival analysis with the log-rank test and univariable and multivariable Cox analyses were conducted to explore survival differences among the subgroups.
Results
Three radiotranscriptomics subtypes (RTSs) were identified based on the radiomics and pathways enrichment profiles. The three RTSs were characterized as having specific molecular hallmarks: RTS1 (proliferation subtype), RTS2 (metabolism subtype), and RTS3 (immune activation subtype). RTS3 showed increased infiltration of most immune cells. The RTS stratification strategy was validated in a validation cohort and showed significant prognostic value. Survival analysis demonstrated that the RTS strategy could stratify NSCLC patients according to prognosis (p = 0.009), and the RTS strategy remained an independent prognostic indicator after adjusting for other clinical parameters.
Conclusions
This radiotranscriptomics study provides a stratification strategy for NSCLC that could provide information for radiomics feature molecular annotation and prognostic prediction.
Key Points
• Radiotranscriptomics subtypes (RTSs) could be used to stratify molecularly heterogeneous patients.
• RTSs showed relationships between molecular phenotypes and radiomics features.
• The RTS algorithm could be used to identify patients with poor prognosis.






Similar content being viewed by others
Abbreviations
- CI:
-
Confidence interval
- DICOM:
-
Digital Imaging and Communications in Medicine
- FPKM:
-
Fragments per kilobase of transcript per million mapped reads
- GLCM:
-
Gray level cooccurrence matrix
- GLDM:
-
Gray level dependence matrix
- GLRLM:
-
Gray level run length matrix
- GLSZM:
-
Gray level size zone matrix
- GSVA:
-
Gene Set Variation Analysis
- HR:
-
Hazard ratio
- K-M:
-
Kaplan-Meier
- NGTDM:
-
Neighbouring gray tone difference matrix
- NSCLC:
-
Non-small cell lung cancer
- ROI:
-
Region of interest
- RTS:
-
Radiotranscriptomics subtype
- TCIA:
-
The Cancer Imaging Archive
- UMAP:
-
Uniform manifold approximation and projection
References
Bera K, Braman N, Gupta A, Velcheti V, Madabhushi A (2022) Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat Rev Clin Oncol 19:132–146
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
Dercle L, Zhao B, Gonen M et al (2022) Early readout on overall survival of patients with melanoma treated with immunotherapy using a novel imaging analysis. JAMA Oncol 8:385–392
Xu X, Zhang HL, Liu QP et al (2019) Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J Hepatol 70:1133–1144
Sun R, Limkin EJ, Vakalopoulou M et al (2018) A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 19:1180–1191
Xi IL, Zhao Y, Wang R et al (2020) Deep learning to distinguish benign from malignant renal lesions based on routine MR imaging. Clin Cancer Res 26:1944–1952
Feng S, Xia T, Ge Y et al (2022) Computed tomography imaging-based radiogenomics analysis reveals hypoxia patterns and immunological characteristics in ovarian cancer. Front Immunol 13:868067
An J, Oh M, Kim SY et al (2022) PET-based radiogenomics supports mTOR pathway targeting for hepatocellular carcinoma. Clin Cancer Res 28:1821–1831
Andre F, Ismaila N, Allison KH et al (2022) Biomarkers for adjuvant endocrine and chemotherapy in early-stage breast cancer: ASCO guideline update. J Clin Oncol 40:1816–1837
Wolf DM, Yau C, Wulfkuhle J et al (2022) Redefining breast cancer subtypes to guide treatment prioritization and maximize response: predictive biomarkers across 10 cancer therapies. Cancer Cell 40(609–623):e606
Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS (2021) Lung cancer. Lancet 398:535–554
Shiri I, Amini M, Nazari M et al (2022) Impact of feature harmonization on radiogenomics analysis: prediction of EGFR and KRAS mutations from non-small cell lung cancer PET/CT images. Comput Biol Med 142:105230
Yamazaki M, Yagi T, Tominaga M, Minato K, Ishikawa H (2022) Role of intratumoral and peritumoral CT radiomics for the prediction of EGFR gene mutation in primary lung cancer. Br J Radiol 95:20220374
Nair JKR, Saeed UA, McDougall CC et al (2021) Radiogenomic models using machine learning techniques to predict EGFR mutations in non-small cell lung cancer. Can Assoc Radiol J 72:109–119
Clark K, Vendt B, Smith K et al (2013) The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26:1045–1057
Aerts HJ, Velazquez ER, Leijenaar RT et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Aerts HJWL, Rios Velazquez E, Leijenaar RTH et al (2015) Data From NSCLC-radiomics-genomics. Cancer Imaging Arch. https://doi.org/10.7937/K9/TCIA.2015.L4FRET6Z
Bakr S, Gevaert O, Echegaray S et al (2018) A radiogenomic dataset of non-small cell lung cancer. Sci Data 5:180202
Bakr S, Olivier G, Echegaray S et al (2017) Data for NSCLC radiogenomics collection. Cancer Imaging Arch. https://doi.org/10.7937/K9/TCIA.2017.7hs46erv
Aerts HJWL, Velazquez ER, Leijenaar RTH et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. https://doi.org/10.1038/ncomms5006(link)
Gevaert O, Xu J, Hoang CD et al (2012) Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data–methods and preliminary results. Radiology 264:387–396
Ji GW, Zhu FP, Xu Q et al (2020) Radiomic features at contrast-enhanced CT predict recurrence in early stage hepatocellular carcinoma: a multi-institutional study. Radiology 294:568–579
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Charoentong P, Finotello F, Angelova M et al (2017) Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 18:248–262
Xu T, Le TD, Liu L et al (2017) CancerSubtypes: an R/Bioconductor package for molecular cancer subtype identification, validation and visualization. Bioinformatics 33:3131–3133
Wang C, Ma J, Shao J et al (2022) Non-invasive measurement using deep learning algorithm based on multi-source features fusion to predict PD-L1 expression and survival in NSCLC. Front Immunol 13:828560
Wong CW, Chaudhry A (2020) Radiogenomics of lung cancer. J Thorac Dis 12:5104–5109
Rossi G, Barabino E, Fedeli A et al (2021) Radiomic detection of EGFR mutations in NSCLC. Cancer Res 81:724–731
Perez-Johnston R, Araujo-Filho JA, Connolly JG et al (2022) CT-based radiogenomic analysis of clinical stage I lung adenocarcinoma with histopathologic features and oncologic outcomes. Radiology 303:664–672
Tu W, Sun G, Fan L et al (2019) Radiomics signature: a potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer 132:28–35
Liu D, Chen J, Ge H et al (2022) Radiogenomics to characterize the immune-related prognostic signature associated with biological functions in glioblastoma. Eur Radiol. https://doi.org/10.1007/s00330-022-09012-x
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Boehm KM, Aherne EA, Ellenson L et al (2022) Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer. Nat Cancer 3:723–733
Feng L, Liu Z, Li C et al (2022) Development and validation of a radiopathomics model to predict pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer: a multicentre observational study. Lancet Digit Health 4:e8–e17
Acknowledgements
The authors acknowledge The Cancer Imaging Archive (TCIA) and Gene Expression Omnibus (GEO) databases for sharing their dataset publicly. The authors also thank the guidance of professor Sandy Napel and professor Leonard Wee.
Funding
This study has received funding by Guangxi Natural Science Foundation under Grant (No. 2020GXNSFDA238005) and Innovation Project of Guangxi Graduate Education (No. YCBZ2022077).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Hong Yang.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional Review Board approval was obtained.
Study subjects or cohorts overlap
Some study subjects or cohorts have been previously reported in Aerts HJ et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006; Bakr S et al. A radiogenomic dataset of non-small cell lung cancer. Sci Data 5:180202.
Methodology
• retrospective
• observational
• multicenter study
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lin, P., Lin, Yq., Gao, Rz. et al. Integrative radiomics and transcriptomics analyses reveal subtype characterization of non-small cell lung cancer. Eur Radiol 33, 6414–6425 (2023). https://doi.org/10.1007/s00330-023-09503-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-023-09503-5