Abstract
Objectives
Radiomics of soft tissue sarcomas (STS) is assumed to correlate with histologic and molecular tumor features, but radiogenomics analyses are lacking. Our aim was to identify if distinct patterns of natural evolution of STS obtained from consecutive pre-treatment MRIs are associated with differential gene expression (DGE) profiling in a pathway analysis.
Methods
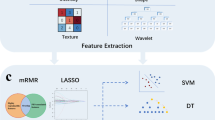
All patients with newly diagnosed STS treated in a curative intent in our sarcoma reference center between 2008 and 2019 and with two available pre-treatment contrast-enhanced MRIs were included in this retrospective study. Radiomics features (RFs) were extracted from fat-sat contrast-enhanced T1-weighted imaging. Log ratio and relative change in RFs were calculated and used to determine grouping of samples based on a consensus hierarchical clustering. DGE and oncogenesis pathway analysis were performed in the delta-radiomics groups identified in order to detect associations between delta-radiomics patterns and transcriptomics features of STS. Secondarily, the prognostic value of the delta-radiomics groups was investigated.
Results
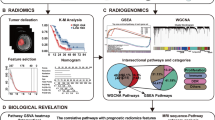
Sixty-three patients were included (median age: 63 years, interquartile range: 52.5–70). The consensus clustering identified 3 reliable delta-radiomics patient groups (A, B, and C). On imaging, group B patients were characterized by increase in tumor heterogeneity, necrotic signal, infiltrative margins, peritumoral edema, and peritumoral enhancement before the treatment start (p value range: 0.0019–0.0244), and, molecularly, by downregulation of natural killer cell–mediated cytotoxicity genes and upregulation of Hedgehog and Hippo signaling pathways. Group A patients were characterized by morphological stability of pre-treatment MRI traits and no local relapse (log-rank p = 0.0277).
Conclusions
This study highlights radiomics and transcriptomics convergence in STS. Proliferation and immune response inhibition were hyper-activated in the STS that were the most evolving on consecutive imaging.
Key Points
• Three consensual and stable delta-radiomics clusters were identified and captured the natural patterns of morphological evolution of STS on pre-treatment MRIs.
• These 3 patterns were explainable and correlated with different well-known semantic radiological features with an ascending gradient of pejorative characteristics from the A group to C group to B group.
• Gene expression profiling stressed distinct patterns of up/downregulated oncogenetic pathways in STS from B group in keeping with its most aggressive radiological evolution.






Similar content being viewed by others
Abbreviations
- AC:
-
Absolute change
- CE:
-
Contrast-enhanced
- CI:
-
Confidence interval
- DGE:
-
Differential gene expression
- FNCLCC:
-
French “Fédération Nationale des Centres de Lutte Contre le Cancer”
- FS:
-
Fat sat
- HR:
-
Hazard ratio
- IQR:
-
Interquartile range
- IRB:
-
Institutional Review Board
- LD:
-
Longest diameter
- LFS:
-
Local relapse-free survival
- LR:
-
Log ratio
- MFS:
-
Metastatic relapse-free survival
- MRI:
-
Magnetic resonance imaging
- NK:
-
Natural killer
- OS:
-
Overall survival
- RF:
-
Radiomics feature
- RNA:
-
Ribonucleic acids
- SAM:
-
Significance analysis of microarrays
- SI:
-
Signal intensity
- STS:
-
Soft tissue sarcoma
- TSE:
-
Turbo spin echo
- TWAC:
-
Time-weighted absolute change
- TWRC:
-
Time-weighted relative change
- WI:
-
Weighted imaging
References
Fletcher CDMF, Bridge JA, Hogendoorn PCW, Martens F (2020) WHO classification of soft tissue and bone tumours, 5th edn. IARC Publications, Lyon, France
Soomers V, Husson O, Young R et al (2020) The sarcoma diagnostic interval: a systematic review on length, contributing factors and patient outcomes. ESMO Open 5:e000592
Gronchi A, Miah AB, Dei Tos AP et al (2021) Soft tissue and visceral sarcomas: ESMO-EURACAN-GENTURIS Clinical Practice Guidelines for diagnosis, treatment and follow-up☆. Ann Oncol S0923-7534(21):02184–0
Callegaro D, Miceli R, Bonvalot S et al (2016) Development and external validation of two nomograms to predict overall survival and occurrence of distant metastases in adults after surgical resection of localised soft-tissue sarcomas of the extremities: a retrospective analysis. Lancet Oncol 17:671–680
Coindre JM, Terrier P, Bui NB et al (1996) Prognostic factors in adult patients with locally controlled soft tissue sarcoma. A study of 546 patients from the French Federation of Cancer Centers Sarcoma Group. J Clin Oncol 14:869–877
Zhao F, Ahlawat S, Farahani SJ et al (2014) Can MR imaging be used to predict tumor grade in soft-tissue sarcoma? Radiology 272:192–201
Crombé A, Marcellin P-J, Buy X et al (2019) Soft-tissue sarcomas: assessment of MRI features correlating with histologic grade and patient outcome. Radiology 291:710–721
Peeken JC, Neumann J, Asadpour R et al (2021) Prognostic assessment in high-grade soft-tissue sarcoma patients: a comparison of semantic image analysis and radiomics. Cancers (Basel) 13:1929
Limkin EJ, Sun R, Dercle L et al (2017) Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol 28:1191–1206
Lambin P, Leijenaar RTH, Deist TM, et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762
Corino VDA, Montin E, Messina A et al (2018) Radiomic analysis of soft tissues sarcomas can distinguish intermediate from high-grade lesions. J Magn Reson Imaging 47:829–840
Peeken JC, Spraker MB, Knebel C et al (2019) Tumor grading of soft tissue sarcomas using MRI-based radiomics. EBioMedicine 48:332–340
Yan R, Hao D, Li J et al (2021) Magnetic resonance imaging-based radiomics nomogram for prediction of the histopathological grade of soft tissue sarcomas: a two-center study. J Magn Reson Imaging 53:1683–1696
Wang H, Chen H, Duan S et al (2020) Radiomics and machine learning with multiparametric preoperative MRI may accurately predict the histopathological grades of soft tissue sarcomas. J Magn Reson Imaging 51:791–797
Crombé A, Fadli D, Buy X et al (2020) High-grade soft-tissue sarcomas: can optimizing dynamic contrast-enhanced MRI postprocessing improve prognostic radiomics models? J Magn Reson Imaging 52:282–297
Crombé A, Le Loarer F, Sitbon M et al (2020) Can radiomics improve the prediction of metastatic relapse of myxoid/round cell liposarcomas? Eur Radiol 30:2413–2424
Spraker MB, Wootton LS, Hippe DS et al (2019) MRI radiomic features are independently associated with overall survival in soft tissue sarcoma. Adv Radiat Oncol 4:413–421
Crombé A, Périer C, Kind M et al (2019) T2 -based MRI delta-radiomics improve response prediction in soft-tissue sarcomas treated by neoadjuvant chemotherapy. J Magn Reson Imaging 50:497–510
Peeken JC, Asadpour R, Specht K et al (2021) MRI-based delta-radiomics predicts pathologic complete response in high-grade soft-tissue sarcoma patients treated with neoadjuvant therapy. Radiother Oncol 164:73–82
Chen S, Li N, Tang Y et al (2021) Radiomics analysis of fat-saturated T2-weighted MRI sequences for the prediction of prognosis in soft tissue sarcoma of the extremities and trunk treated with neoadjuvant radiotherapy. Front Oncol 11:710649
Fadli D, Kind M, Michot A et al (2021) Natural changes in radiological and radiomics features on MRIs of soft-tissue sarcomas naïve of treatment: correlations with histology and patients’ outcomes. J Magn Reson Imaging. https://doi.org/10.1002/jmri.28021
Trojani M, Contesso G, Coindre JM et al (1984) Soft-tissue sarcomas of adults; study of pathological prognostic variables and definition of a histopathological grading system. Int J Cancer 33:37–42
McCormick M, Liu X, Jomier J et al (2014) ITK: enabling reproducible research and open science. Front Neuroinform 8:13
Tustison NJ, Avants BB, Cook PA et al (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320
Nyúl LG, Udupa JK (1999) On standardizing the MR image intensity scale. Magn Reson Med 42:1072–1081
Nakamura T, Matsumine A, Matsubara T et al (2017) Infiltrative tumor growth patterns on magnetic resonance imaging associated with systemic inflammation and oncological outcome in patients with high-grade soft-tissue sarcoma. PLoS One 12:e0181787
Law CW, Chen Y, Shi W, Smyth GK (2014) voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29.
Wilkerson MD, Hayes DN (2010) ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26:1572–1573
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98:5116–5121
Subramanian A, Tamayo P, Mootha VK et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550
Newman AM, Liu CL, Green MR et al (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12:453–457
Hanna SL, Fletcher BD, Parham DM, Bugg MF (1991) Muscle edema in musculoskeletal tumors: MR imaging characteristics and clinical significance. J Magn Reson Imaging 1:441–449
Crombé A, Le Loarer F, Stoeckle E et al (2018) MRI assessment of surrounding tissues in soft-tissue sarcoma during neoadjuvant chemotherapy can help predicting response and prognosis. Eur J Radiol 109:178–187
White LM, Wunder JS, Bell RS et al (2005) Histologic assessment of peritumoral edema in soft tissue sarcoma. Int J Radiat Oncol Biol Phys 61:1439–1445
Fortes-Andrade T, Almeida JS, Sousa LM et al (2021) The role of natural killer cells in soft tissue sarcoma: prospects for immunotherapy. Cancers (Basel) 13:3865
Kelleher FC, Cain JE, Healy JM et al (2012) Prevailing importance of the hedgehog signaling pathway and the potential for treatment advancement in sarcoma. Pharmacol Ther 136:153–168
Ahlawat S, Fritz J, Morris CD, Fayad LM (2019) Magnetic resonance imaging biomarkers in musculoskeletal soft tissue tumors: review of conventional features and focus on nonmorphologic imaging. J Magn Reson Imaging 50:11–27
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Prof. Antoine Italiano.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
Three of the authors have significant statistical expertise (C.L. and F.B. are bioinformaticians; A.C. has a PhD in applied mathematics).
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Study subjects or cohorts overlap
Some study subjects or cohorts have been previously reported in the study by Fadli et al [20].
Methodology
• retrospective
• diagnostic or prognostic study/observational/experimental
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Crombé, A., Bertolo, F., Fadli, D. et al. Distinct patterns of the natural evolution of soft tissue sarcomas on pre-treatment MRIs captured with delta-radiomics correlate with gene expression profiles. Eur Radiol 33, 1205–1218 (2023). https://doi.org/10.1007/s00330-022-09104-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-09104-8