Abstract
Objectives
Relapse occurs in ~20% of patients with classical Hodgkin lymphoma (cHL) despite treatment adaption based on 2-deoxy-2-[18F]fluoro-d-glucose positron emission tomography/computed tomography response. The objective was to evaluate pre-treatment FDG PET/CT–derived machine learning (ML) models for predicting outcome in patients with cHL.
Methods
All cHL patients undergoing pre-treatment PET/CT at our institution between 2008 and 2018 were retrospectively identified. A 1.5 × mean liver standardised uptake value (SUV) and a fixed 4.0 SUV threshold were used to segment PET/CT data. Feature extraction was performed using PyRadiomics with ComBat harmonisation. Training (80%) and test (20%) cohorts stratified around 2-year event-free survival (EFS), age, sex, ethnicity and disease stage were defined. Seven ML models were trained and hyperparameters tuned using stratified 5-fold cross-validation. Area under the curve (AUC) from receiver operator characteristic analysis was used to assess performance.
Results
A total of 289 patients (153 males), median age 36 (range 16–88 years), were included. There was no significant difference between training (n = 231) and test cohorts (n = 58) (p value > 0.05). A ridge regression model using a 1.5 × mean liver SUV segmentation had the highest performance, with mean training, validation and test AUCs of 0.82 ± 0.002, 0.79 ± 0.01 and 0.81 ± 0.12. However, there was no significant difference between a logistic model derived from metabolic tumour volume and clinical features or the highest performing radiomic model.
Conclusions
Outcome prediction using pre-treatment FDG PET/CT–derived ML models is feasible in cHL patients. Further work is needed to determine optimum predictive thresholds for clinical use.
Key points
• A fixed threshold segmentation method led to more robust radiomic features.
• A radiomic-based model for predicting 2-year event-free survival in classical Hodgkin lymphoma patients is feasible.
• A predictive model based on ridge regression was the best performing model on our dataset.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hodgkin’s lymphoma (HL) is a haematopoietic malignancy characterised by the presence of Reed-Sternberg cells [1]. There are five different sub-classes of HL: nodular lymphocyte-predominant HL (NLPHL), and four under the umbrella category of classical HL (cHL): nodular sclerosing, mixed cellularity, lymphocyte-rich and lymphocyte-depleted. Ninety percent of HL cases are cHL [2]. NLPHL is often treated differently to cHL and is associated with more indolent progression [2]. Given the higher proportion of cHL cases, difference in treatment regimens and higher relapse rate in cHL compared to NLPHL, this paper will focus on cHL only [3].
Chemotherapy is the mainstay of frontline treatment of cHL; the most common regimes being doxorubicin (Adriamycin), bleomycin, vinblastine and dacarbazine (ABVD), or bleomycin, etoposide, doxorubicin (Adriamycin), cyclophosphamide, vincristine (Oncovin), procarbazine, and prednisone (BEACOPP) [4]. The treatment regime and number of cycles can vary depending on patient risk factors, disease stage and initial treatment response. Radiotherapy is used in patients with stage 1 or localised stage 2 disease or in residual bulky disease [4]. The gold standard imaging modality for staging and response assessment in HL is 2-deoxy-2-[18F]fluoro-d-glucose (FDG) positron emission tomography/computed tomography (PET/CT) [5]. Patients typically undergo PET/CT pre-treatment, following two cycles of chemotherapy (interim) and post-treatment. Interim PET/CT is used to guide treatment adaption, balancing the risk of chemotherapy-associated toxicity with maximising chances of event-free survival (EFS) [6]. Five-year survival in HL is approximately 86% [7]. However, even following complete metabolic response (CMR), approximately 20% of cHL patients will relapse with 72% of relapses occurring within the first 2 years of diagnosis [8]. The ability to identify patients at greater risk of relapse pre-treatment would allow upfront treatment stratification and could improve outcomes.
Previous studies assessing imaging parameters derived from baseline PET/CT for outcome prediction have mainly focused on metabolic tumour volume (MTV), total lesion glycolysis (TLG) and maximum or mean standardised uptake value (SUVmax and SUVmean) [9]. SUV is defined as the ratio of injected radioactivity within an image at a given timepoint when compared to the whole-body [10]. MTV is the volume of metabolically active segmented disease, with different segmentation techniques described [11]. The TLG is MTV multiplied by the SUVmean. Radiomics transforms images into mineable high-dimensional data permitting invisible feature extraction, analysis and modelling [12]. A limited number of studies using small sample sizes have demonstrated the potential of radiomic features in predicting progression-free survival (PFS) or overall survival (OS) in HL patients [13,14,15,16]. The aim of this work was to evaluate the performance of models using radiomic features derived from pre-treatment FDG PET/CT to predict 2-year EFS in cHL patients using a larger tertiary centre cohort of patients.
Methods
This study adhered to the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) guidelines (Supplemental Material 1).
Patient selection
Retrospective review of radiology and clinical databases was performed to identify patients who had undergone FDG PET/CT for baseline staging of cHL at our institution between January 2008 and January 2018. This was chosen as the cut-off to allow a minimum of 2-year follow-up without confounding factors introduced by the COVID-19 pandemic. Patients were excluded if they were under 16 years of age, did not have cHL, had treatment prior to their staging PET/CT study, did not have measurable disease on PET/CT, had a concurrent malignancy or if the images were degraded or incomplete. Patients who had hepatic disease or had no measurable disease above 4.0 SUV were removed as this would influence the segmentation techniques used.
Patient age, ethnicity, disease stage, date of PET/CT, scanner model and protocol used, type and length of treatment, date of recurrence (confirmed by imaging or clinical examination), last clinical contact and length of follow-up or date of death were all recorded from electronic notes, from radiological records and from a regional haematological malignancy database. An event was defined as relapse, recurrence or death within the 2-year follow-up period. Due to missing clinical data, it was not possible to evaluate scoring systems such as the international prognostic score.
Informed written consent was obtained prospectively from all patients at the time of imaging for use of anonymised images in research and service development projects. As this was a retrospective study, not involving patient contact or the alteration of treatment, following discussion with the Research and Innovation Department at LTHT, it was agreed that this represented a service improvement project and was approved by the University of Leeds School of Medicine Research Ethics Committee (SoMREC).
PET/CT acquisition
PET/CT studies were performed as part of routine clinical care using a standardised protocol. All patients fasted for 6h prior to administration of intravenous FDG (4MBq/kg). If serum blood glucose was > 10 mmol/L, the study was rescheduled following a clinical review of the patient’s diabetic control. Patients were scanned 1 h following FDG administration. Scans were acquired using a 16-slice Discovery STE PET/CT scanner (GE Healthcare) prior to June 2010; a 64-slice Philips Gemini TF64 scanner (Philips Healthcare) between June 2010 and October 2015; and 64-slice Discovery 690 or 710 scanners (GE Healthcare) after October 2015 (Table 1). Attenuation correction was performed using a CT component acquired with the following settings: 140 kV; 80 mAs; pitch 6; 3.75-mm slice thickness.
Image segmentation, feature extraction and machine learning analysis
A detailed methodology including detail of who performed the segmentation and interpretation of images is available in Supplemental Material 2. Two semi-automated segmentation techniques were used to contour the total lymphomatous disease within each study: the first using a fixed threshold of 4.0 SUV, and the second using a threshold of 1.5 × liver SUVmean. This method has been used in different cancer types (RTx v1.8.2, Mirada Medical) [17, 18]. Ten percent of cases were re-segmented using the same methodology following a 3-month washout period using Slicer (v4.11). These re-segmentations were used to investigate the robustness of the extracted radiomic features using different bin widths/bin numbers. Both the CT and PET images were resampled to a uniform voxel of 2 mm3. Features were extracted using PyRadiomics (v2.2.0) with 3935 features (PET/CT component × (shape features + first and second order features × number of filters)) extracted per segmentation technique for each patient (Supplemental Material 2: Table 1). Harmonisation to account for the different scanners was applied using the ComBat method (https://github.com/Jfortin1/ComBatHarmonization) [19].
The data was split into training and test cohorts stratified around 2-year EFS (2-EFS), age, sex, ethnicity, disease stage, having radiotherapy, having ABVD-based chemotherapy and being treated as advanced disease using scikit-learn (v0.24.2). The cohorts were split using an 80:20 ratio. Mann-Whitney U and χ2 tests (SciPy v1.6.3) were used to assess for significance in continuous and categorical clinical characteristics between the training and test cohorts respectively. A p value less than 0.05 was regarded as significant. Correlated features were removed if the Pearson coefficient was over 0.8. Seven different machine learning methods were used to create prediction models (scikit-learn v0.24.2): random forest, logistic regression (elastic net, lasso and ridge penalties explored), k-nearest neighbour (KNN), single-layer perceptron (SLP), multi-layer perceptron (MLP), Gaussian process classifier (GCP) and support vector machine (SVM). A maximum number of five features were selected for each of these models. The features selected in each method are based on the highest mean receiver operating characteristic (ROC) area under the curve (AUC) in five-fold stratified cross-validation with 20 repeats.
Each model was trained and tuned on the training cohort, using a five-fold cross-validation stratified around 2-EFS, again with 20 repeats. The model, hyperparameter and feature selection combination with the highest mean validation score from both the 4.0 SUV and 1.5 × mean liver segmentation were tested once on the unseen test cohort data. Given the growing literature surrounding the use of MTV as an outcome predictor, a separate logistic regression model using total MTV was trained in addition to a model using only clinical features and a combined clinical and MTV model. AUCs were compared using the DeLong method [20]. An appropriate threshold from the ROC curve for each of the best performing models was derived using the Youden index with the Matthews correlation coefficient (MCC), sensitivity, specificity, positive predictive value (PPV) and negative predictive value (PPV) presented.
Results
Patient demographics
A total of 289 patients were included in the study, with the patient demographics detailed in Table 2. There were no significant differences in the clinical characteristics between training and test cohorts.
Bin widths
For both the 4.0 SUV and 1.5 × mean liver SUV segmentation techniques, bin widths for PET and CT data were most robust when derived from the maximum range of SUV or HU respectively divided by 128 (Supplementary Figures 1 and 2). Overall, the 4.0 SUV segmentation technique resulted in more radiomic features being robust than the 1.5 × mean liver SUV segmentation method.
Clinical- and MTV-derived models of 2-EFS
Patients who had a 2-EFS event had a significantly larger MTV compared to those who did not have a 2-EFS event. This was true for both segmentation techniques. With the 4.0 SUV method, the median MTVs were 167.4 cm3 versus 87.9 cm3 (p = 0.03); and for the 1.5 × mean liver SUV method, 324.3 cm3 versus 148.6 cm3 (p = 0.009). The median volumes were significantly greater in patients treated as advanced disease. For the 4.0 SUV method, the median MTVs were 250.6 cm3 (2-EFS event) versus 110.4 cm3 (no event) (p = 0.03); and for the 1.5 × mean liver SUV method, 457.8 cm3 (2-EFS event) versus 227.9 cm3 (no event) (p = 0.02)
A logistic regression model using MTV derived from a 4.0 SUV method resulted in a mean training AUC of 0.61 ± 0.02 (mean ± 95% CI) and a mean validation AUC of 0.61 ± 0.10 with the odds ratio being 1.00038 (Table 3). The logistic regression model derived from MTV using the 1.5 × mean liver SUV method had a mean training AUC of 0.63 ± 0.02 and a mean validation AUC of 0.63 ± 0.10, with the odds ratio being 1.00038.
Cancer stage 1, cancer stage 4 and age were selected as features for the clinical-based logistic regression model. This had a mean training AUC of 0.74 ± 0.004 and a mean validation AUC of 0.74 ± 0.02. When combing the features from this model with 1.5 × mean liver SUV MTV, the model had a mean training AUC of 0.74 ± 0.004 and a mean validation AUC of 0.72 ± 0.01. This model was tested on the unseen test set and achieved an AUC of 0.68 ± 0.11 (Fig. 1), MCC of 0.27, sensitivity of 0.31, specificity of 0.91, NPV of 0.47 and PPV of 0.85 at a threshold of 0.45.
Clinical and radiomic model for the prediction of 2-EFS
The predictive model with the highest AUC was a ridge regression model derived from clinical and radiomic features extracted from the 1.5 × mean SUV threshold segmentation technique (Table 4). The model was constructed using features selected using a forward wrapper with five features chosen. The hyperparameters of the model were as follows: C = 1, penalty = l2 and solver = sag, class weight = balanced. The features chosen were age, PET flatness, PET major axis length, PET logarithm GLSZM size zone non-uniformity normalised, PET lbp-3D-m1 GLCM correlation and PET lbp-3D-m2 first order skewness. The mean training AUC was 0.82 ± 0.002, the mean validation AUC was 0.79 ± 0.01 and the test AUC was 0.81 ± 0.12 (Fig. 2), with MCC = 0.43, sensitivity = 0.42, specificity = 0.94, NPV = 0.67 and PPV = 0.85. The demographics of the mislabelled patients are presented in Supplementary Material 2: Table 2.
The highest performing predictive model using the 4.0 SUV threshold was a regression model using a ridge regression penalty with a mean training AUC of 0.79 ± 0.002, the mean validation AUC of 0.77 ± 0.01 and the test AUC of 0.74 ± 0.13 (Fig. 3). The MCC = 0.30, sensitivity = 032, specificity = 0.95, NPV = 0.42 and PPV = 0.92 at a threshold of 0.27. The model was constructed using features selected from a forward wrapper method of feature selection with five features chosen. The hyperparameters of the model were as follows: C = 100, penalty = l2 and solver = saga, class weight = balanced.
There was no significant difference between the test set AUCs of the best performing clinical- and radiomic-based models with each other and with the best performing clinical- and MTV-based model (Fig. 4; Table 5). The intercept and coefficients for each model are presented in Supplementary Material 2: Table 3.
Discussion
This study confirms that pre-treatment outcome prediction using FDG PET/CT–derived radiomic features is feasible in patients with cHL. The best performing model was created using ridge regression combining age and four radiomic features (PET flatness, PET major axis length, PET logarithm GLSZM size zone non-uniformity normalised, PET lbp-3D-m1 GLCM correlation and PET lbp-3D-m2 first order skewness) extracted from PET images using a 1.5 × mean liver SUV method with a bin width of 0.24. It must be noted that there was no significant difference between the test AUC of this model and those of a combined clinical and MTV model and a model created using 4.0 SUV fixed threshold segmentation. This is likely due to small numbers involved given the relatively large confidence intervals. Due to missing clinical data, it was not possible to adjust for features used to stratify patients into early and advanced disease. A surrogate, treatment intent, was used instead which demonstrated that the models created remained reasonable predictors of outcome for patients treated as having advanced disease.
Further work should be performed to assess the relationship of ethnicity and socio-economic status on a model’s predictive ability to avoid creation of a model which discriminates against under-represented subsets of patients due to lack of data to train and test the model on [21, 22]. Unadjusted confounders are likely one of the reasons for a minority of studies reporting the poor ability of MTV as an outcome predictor in lymphoma [23,24,25]. Most notably Adams et al found that MTV was not an independent predictor of overall survival or PFS in diffuse large B cell lymphoma once adjusting for the National Comprehensive Cancer Network International Prognostic Index [26]. To allow for transparency, our study has provided the demographic information for the patients who were mislabelled using the predictive model with the highest test AUC.
Two different segmentation techniques were explored. The first was a fixed threshold of 4.0 SUV which has been demonstrated to be a reproducible, efficient method for contouring disease [27]. The second was 1.5 × mean liver SUV which has been explored in other malignancies and provides an adaptive threshold which adjusts for background SUV uptake [17, 18]. Our study echoed previous work demonstrating a fixed threshold led to more features being robust following re-segmentation. The fixed thresholding segmentation technique required less steps, and less manual adaption [28]. However, a fixed SUV thresholding technique does not scale with the physiological uptake and therefore, the contours may vary on repeat studies due to external effects on the SUV rather than tumour pathophysiology [27]. The study also demonstrated the variability which can occur when repeating a segmentation methodology on different software (Fig. 5), with radiomic features not being deemed robust following repeated segmentation even when using the same SUV thresholds. ComBat harmonisation was employed to mitigate against the effects of scanner variation. This is based on Bayes theorem and attempts to predict scanner influence whilst maintaining biological variation [29]. For this to be effective, however, there must be enough samples from different scanners to apply the harmonisation method [30] and it cannot be applied prospectively to scanner acquisitions outside those used for training of the predictive model.
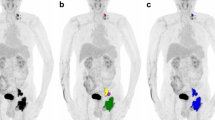
Select axial slice through PET (a) and CT (b) images of a patient with Hodgkin lymphoma demonstrating a pathological left level II lymph node. The purple segmentation represents the original 4.0 SUV fixed threshold segmentation performed using Mirada Medical RTx (v1.8.2) software and the green areas represent the additional area included when segmented with a fixed 4.0 SUV threshold using 3D Slicer (v4.11) software
Previous studies have explored the use of radiomic features in the prediction of outcomes in HL [13,14,15]. Lue et al found SUV kurtosis, stage and intensity non-uniformity (INU) derived from Grey Level Run Length Matrix (GLRLM) were independent predictors of PFS in a small cohort of 42 patients. Milogrom et al demonstrated that the combination of SUVmax, MTV, InformationMeasureCorr1, InformationMeasureCorr2 and InverseVariance derived from GLCM 2.5 had an AUC of 0.95 when predicting relapse in 167 patients with stage I–II HL. However, there were very few events, with the validation cohort only having two patients who relapsed. Sollini et al assessed a radiomic fingerprint using principal component analysis to classify patients who would relapse within 4 years of treatment in a cohort of 85 patients. They explored fingerprints created from a single largest nodal or extra-nodal lesion versus using all lesions and found that the intra-patient similarity was low, and that the highest accuracy was achieved when using all lesions within the model [15]. This highlights the inherent heterogeneity of radiomic features within different lesions and that by restricting analysis to a single lesion, the predictive model may also be limited. The current study of 289 patients is one of the largest to assess potential utility of radiomic features derived from pre-treatment FDG PET/CT for predicting outcome in cHL patients. It demonstrates that radiomics could feasibly improve prediction of 2-EFS. However, this requires validation on an independent external dataset and although the AUC for the test set was 0.81, no clear predictive threshold could be derived. This must be a key target when creating any machine learning or AI-based model. In terms of HL, it would be the ability to balance side effects of escalated treatment, with the rates of EFS and toxicity varying between treatment regimens [31]. The advent of newer therapeutic strategies limits the use of predictive models made on retrospective data; future efforts should focus on validating imaging, genetic and clinical predictive features in carefully designed prospective, multi-centre clinical trials.
A TRIPOD checklist was used to ensure transparency of the study’s methodology, a concern in previous radiomic studies [32, 33]. However, no external validation was performed, and although contouring was undertaken without knowledge of clinical outcome, no measures to blind assessors were specifically undertaken. Although patients with other concurrent malignancies were excluded from analysis, other pathologies were not taken into consideration when looking at mortality. Other study limitations include its retrospective nature, the relatively small event rate, reliance on clinical records to determine date of relapse/recurrence, exclusion of patients with hepatic disease/or without disease > 4.0 SUV and variation in different patient’s treatment regimen.
Conclusion
There is potential for models derived from radiomic features extracted from pre-treatment FDG PET/CT to predict 2-EFS in cHL patients. Further work is needed to determine optimum thresholds for clinical use.
Abbreviations
- ABVD:
-
Doxorubicin (Adriamycin), bleomycin, vinblastine and dacarbazine
- AUC:
-
Area under the curve
- BEACOPP:
-
Bleomycin, etoposide, doxorubicin (Adriamycin), cyclophosphamide, vincristine (Oncovin), procarbazine, and prednisone
- cHL:
-
Classical Hodgkin lymphoma
- CMR:
-
Complete metabolic response
- ComBat:
-
Combating batch effects when combining batches
- DICOM:
-
Digital imaging and communications in medicine
- EFS:
-
Event-free survival
- FDG:
-
2-deoxy-2-[18F]fluoro-d-glucose
- GLCM:
-
Grey level co-occurrence matrix
- GLDM:
-
Grey level dependence matrix
- GLRLM:
-
Grey level run length matrix
- GLSZM:
-
Grey level size zone matrix
- HL:
-
Hodgkin lymphoma
- HU:
-
Hounsfield units
- Id:
-
Inverse difference
- Idm:
-
Inverse difference moment
- Idmn:
-
Inverse difference moment normalised
- Idn:
-
Inverse difference normalised
- Imc:
-
Informational measure of correlation
- KNN:
-
k-nearest neighbour
- lbfgs:
-
Limited memory Broyden-Fletcher-Goldfarb-Shanno
- lbp:
-
Local binary pattern
- LGLE:
-
Low grey level emphasis selected
- MCC:
-
Matthews correlation coefficient
- MTV:
-
Metabolic tumour volume
- NGTDM:
-
Neighbouring grey tone difference matrix
- NIfTI:
-
Neuroimaging Informatics Technology Initiative
- NLPHL:
-
Nodular lymphocyte-predominant HL
- NPV:
-
Negative predictive value
- PET/CT:
-
Positron emission tomography/computed tomography
- PPV:
-
Positive predictive value
- ROC:
-
Receiver operating characteristic
- ROI:
-
Region of interest
- RT:
-
Radiotherapy
- SUV:
-
Standardised uptake value
- SVM:
-
Support vector machine
- TLG:
-
Total lesion glycolysis
- TRIPOD:
-
Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis
References
Connors JM, Cozen W, Steidl C et al (2020) Hodgkin lymphoma. Nat Rev Dis Prim 6(1):61
Shanbhag S, Ambinder RF (2018) Hodgkin lymphoma: a review and update on recent progress. CA Cancer J Clin 68(2):116–132
Spinner MA, Varma G, Advani RH (2019) Modern principles in the management of nodular lymphocyte-predominant Hodgkin lymphoma. Br J Haematol 184(1):17–29
Follows GA, Ardeshna KM, Barrington SF et al (2014) Guidelines for the first line management of classical Hodgkin lymphoma. Br J Haematol 166(1):34–49
Kanoun S, Rossi C, Casasnovas O (2018) [18F]FDG-PET/CT in Hodgkin lymphoma: current usefulness and perspectives. Cancers (Basel) 10(5):145
El-Galaly TC, Villa D, Gormsen LC, Baech J, Lo A, Cheah CY (2018) FDG-PET/CT in the management of lymphomas: current status and future directions. J Intern Med 284(4):358–376
SEER (2018) SEER cancer statistics review. In: 1975-2016.
Hapgood G, Zheng Y, Sehn LH et al (2016) Evaluation of the risk of relapse in classical Hodgkin lymphoma at event-free survival time points and survival comparison with the general population in British Columbia. J Clin Oncol 34(21):2493–2500
Frood R, Burton C, Tsoumpas C et al (2021) Baseline PET/CT imaging parameters for prediction of treatment outcome in Hodgkin and diffuse large B cell lymphoma: a systematic review. Eur J Nucl Med Mol Imaging 48(10):3198–3220
Kinahan PE, Fletcher JW (2010) Positron emission tomography-computed tomography standardized uptake values in clinical practice and assessing response to therapy. Semin Ultrasound CT MR 31(6):496–505
Im H-J, Bradshaw T, Solaiyappan M, Cho SY (2018) Current methods to define metabolic tumor volume in positron emission tomography: which one is better? Nucl Med Mol Imaging 52(1):5–15
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14(12):749–762
Milgrom SA, Elhalawani H, Lee J et al (2019) A PET radiomics model to predict refractory mediastinal Hodgkin lymphoma. Sci Rep 9(1):1322
Lue K-H, Wu Y-F, Liu S-H et al (2019) Prognostic value of pretreatment radiomic features of 18F-FDG PET in patients with Hodgkin lymphoma. Clin Nucl Med 44(10):E559–E565
Sollini M, Kirienko M, Cavinato L et al (2020) Methodological framework for radiomics applications in Hodgkin’s lymphoma. Eur J Hybrid Imaging 4:9
Zhou Y, Zhu Y, Chen Z, Li J, Sang S, Deng S (2021) Radiomic features of 18F-FDG PET in Hodgkin lymphoma are predictive of outcomes. Contrast Media Mol Imaging 2021:6347404
Brown PJ, Zhong J, Frood R et al (2019) Prediction of outcome in anal squamous cell carcinoma using radiomic feature analysis of pre-treatment FDG PET-CT. Eur J Nucl Med Mol Imaging 46(13):2790–2799
Zhong J, Frood R, Brown P et al (2021) Machine learning-based FDG PET-CT radiomics for outcome prediction in larynx and hypopharynx squamous cell carcinoma. Clin Radiol 76(1):78.e9–78.e17
Fortin J-P, Cullen N, Sheline YI et al (2018) Harmonization of cortical thickness measurements across scanners and sites. Neuroimage 167:104–120
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44(3):837–845
Berkman AM, Andersen CR, Puthenpura V et al (2021) Impact of race, ethnicity, and socioeconomic status over time on the long-term survival of adolescent and young adult Hodgkin lymphoma survivors. Cancer Epidemiol Biomark Prev 30(9):1717–1725
Pahwa P, Karunanayake CP, Spinelli JJ et al (2009) Ethnicity and incidence of Hodgkin lymphoma in Canadian population. BMC Cancer 9:1–9. https://doi.org/10.1186/1471-2407-9-141
Mettler J, Müller H, Voltin CA et al (2019) Metabolic tumor volume for response prediction in advanced-stage Hodgkin lymphoma. J Nucl Med 60(2):207–211
Tseng D, Rachakonda LP, Su Z et al (2012) Interim-treatment quantitative PET parameters predict progression and death among patients with Hodgkin’s disease. Radiat Oncol. https://doi.org/10.1186/1748-717X-7-5
Gallicchio R, Mansueto G, Simeon V et al (2014) F-18 FDG PET/CT quantization parameters as predictors of outcome in patients with diffuse large B-cell lymphoma. Eur J Haematol 92(5):382–389
Adams HJA, de Klerk JMH, Fijnheer R et al (2015) Prognostic superiority of the National Comprehensive Cancer Network International Prognostic Index over pretreatment whole-body volumetric-metabolic FDG-PET/CT metrics in diffuse large B-cell lymphoma. Eur J Haematol 94(6):532–539
Barrington SF, Zwezerijnen BGJC, de Vet HCW et al (2021) Automated segmentation of baseline metabolic total tumor burden in diffuse large B-cell lymphoma: which method is most successful? A study on behalf of the PETRA consortium. J Nucl Med 62(3):332–337
Driessen J, Zwezerijnen GJ, Schöder H et al (2022) The impact of semi-automatic segmentation methods on metabolic tumor volume, intensity and dissemination radiomics in 18 F-FDG PET scans of patients with classical Hodgkin lymphoma. J Nucl Med. https://doi.org/10.2967/jnumed.121.263067
Orlhac F, Boughdad S, Philippe C et al (2018) A postreconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med 59(8):1321–1328
Orlhac F, Eertink JJ, Cottereau AS et al (2022) A guide to ComBat harmonization of imaging biomarkers in multicenter studies. J Nucl Med 63(2):172–179
Lang N, Crump M (2020) PET-adapted approaches to primary therapy for advanced Hodgkin lymphoma. Ther Adv Hematol 11:204062072091449
Park JE, Kim D, Kim HS et al (2020) Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur Radiol 30(1):523–536
Pinto dos Santos D, Dietzel M, Baessler B (2021) A decade of radiomics research: are images really data or just patterns in the noise? Eur Radiol 31(1):2–5
Funding
R Frood, F Gleeson, C Patel and A Scarsbrook received funding from Innovate UK. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Prof. Andrew Scarsbrook.
Conflict of interest
Scarsbrook is an Associate Editor of European Radiology and a member of the Editorial Board for the journal. They have not taken part in the review or selection process of this article.
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• diagnostic or prognostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 540 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Frood, R., Clark, M., Burton, C. et al. Utility of pre-treatment FDG PET/CT–derived machine learning models for outcome prediction in classical Hodgkin lymphoma. Eur Radiol 32, 7237–7247 (2022). https://doi.org/10.1007/s00330-022-09039-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-09039-0








