Abstract
Objectives
Based on germline and somatic mutation profiles, pheochromocytomas and paragangliomas (PPGLs) can be classified into different clusters. We investigated the use of [18F]FDG-PET/CT radiomics, SUVmax and biochemical profile for the identification of the genetic clusters of PPGLs.
Methods
In this single-centre cohort, 40 PPGLs (13 cluster 1, 18 cluster 2, 9 sporadic) were delineated using a 41% adaptive threshold of SUVpeak ([18F]FDG-PET) and manually (low-dose CT; ldCT). Using PyRadiomics, 211 radiomic features were extracted. Stratified 5-fold cross-validation for the identification of the genetic cluster was performed using multinomial logistic regression with dimensionality reduction incorporated per fold. Classification performances of biochemistry, SUVmax and PET(/CT) radiomic models were compared and presented as mean (multiclass) test AUCs over the five folds. Results were validated using a sham experiment, randomly shuffling the outcome labels.
Results
The model with biochemistry only could identify the genetic cluster (multiclass AUC 0.60). The three-factor PET model had the best classification performance (multiclass AUC 0.88). A simplified model with only SUVmax performed almost similarly. Addition of ldCT features and biochemistry decreased the classification performances. All sham AUCs were approximately 0.50.
Conclusion
PET radiomics achieves a better identification of PPGLs compared to biochemistry, SUVmax, ldCT radiomics and combined approaches, especially for the differentiation of sporadic PPGLs. Nevertheless, a model with SUVmax alone might be preferred clinically, weighing model performances against laborious radiomic analysis. The limited added value of radiomics to the overall classification performance for PPGL should be validated in a larger external cohort.
Key Points
• Radiomics derived from [18F]FDG-PET/CT has the potential to improve the identification of the genetic clusters of pheochromocytomas and paragangliomas.
• A simplified model with SUVmax only might be preferred clinically, weighing model performances against the laborious radiomic analysis.
• Cluster 1 and 2 PPGLs generally present distinctive characteristics that can be captured using [18F]FDG-PET imaging. Sporadic PPGLs appear more heterogeneous, frequently resembling cluster 2 PPGLs and occasionally resembling cluster 1 PPGLs.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
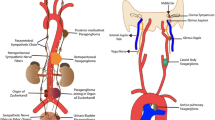
Pheochromocytomas and paragangliomas (PPGLs) are rare catecholamine-producing neuroendocrine tumours arising from the chromaffin cells in the adrenal medulla and extra-adrenal sympathetic ganglia [1]. In around 40% of patients, PPGLs are part of hereditary syndromes caused by germline mutations [2]. Germline mutations in at least a dozen PPGL susceptibility genes with relevant prevalence have been identified, including VHL (von Hippel-Lindau), SDHA/B/C/D/AF2 (succinate dehydrogenase subunits A, B, C, D and assembly factor 2), RET (rearranged during transfection), NF1 (neurofibromatosis type 1), MAX (myc-associated factor X) and TMEM127 (transmembrane protein 127) [3,4,5]. Moreover, in 30–40% of sporadic PPGLs, somatic mutations are identified [5]. Based on both germline and somatic mutations, PPGLs can be classified into two clusters: cluster 1 PPGLs, characterised by increased expression of genes involved in hypoxia (i.e. SDHx/VHL), and cluster 2, associated with kinase signalling (i.e. RET/NF1) [6, 7]. The characterisation of the genetic cluster is of interest, because the underlying mutations directly impact the clinical presentation of PPGLs. The prevalence of malignancy generally varies between 10 and 17%, but reaches even 40% in patients with SDHB mutations [8].
Genetic differences result in phenotypic differences with regard to cellular metabolism, which can be observed in the uptake of 2-[18F]fluoro-2-deoxy-D-glucose ([18F]FDG) by positron emission tomography (PET), with relatively high standardised uptake values (SUV) detected in cluster 1 PPGLs [9, 10]. Previous research in a part of this cohort showed that the maximum SUV (SUVmax) could distinguish hereditary cluster 1 and 2 PPGLs with an area under the receiver operating characteristic curve (AUC) of 0.91 (95% CI: 0.80–1.00) [11]. In addition to the traditional quantitative PET features such as the SUVmax, radiomics allows quantification of tracer uptake heterogeneity and other imaging features [12, 13]. Radiomics, the extraction of large amounts of quantitative features from medical imaging, aims to find stable and clinically relevant image-derived biomarkers that provide a non-invasive way of quantifying and monitoring disease characteristics in clinical practice [14]. Literature on radiomics in PPGLs is scarce, but includes a computed tomography (CT) approach for the differentiation between pheochromocytomas and lipid-poor adenoma [15], an approach on T2 weighted fat-saturated magnetic resonance imaging for the differentiation of paragangliomas from other neck masses [16] and a PET approach studying characterisation of the genetic cluster in pheochromocytomas [17].
This study investigated the potential utility of radiomic features derived from PET and low-dose CT for the characterisation of the genetic cluster of PPGLs.
Materials and methods
Patient population
Patients with PPGL with known mutation status and who underwent a [18F]FDG-PET/CT scan in the Radboud University Medical Center between 2011 and 2018 were retrospectively included. A selection of these patients has previously been studied [10, 11, 18]. This retrospective database study has been reviewed and approved by the Commission on Medical Research Involving Human Subjects Region Arnhem-Nijmegen, the Netherlands. Informed consent was waived because of the retrospective nature of the study. Patients that objected to the use of their anonymised data were excluded.
All patients underwent genetic testing for germline mutations in known susceptibility genes (SDHA/B/C/D/AF2, RET, VHL, TMEM127 and MAX) using standard clinical diagnostic procedures. In case no germline mutation was found, somatic mutations were obtained from post-operative histology. The classes cluster 1 and 2 contain both germline and somatic mutations. The class sporadic contains sporadic PPGLs without known mutations found in germline and tumour tissue associated with cluster 1 or 2. The biochemical diagnosis was based upon the collection of plasma-free metanephrines (metanephrine, normetanephrine and 3-methoxytyramine; metabolites of the catecholamines adrenaline, noradrenaline and dopamine, respectively) and assayed using high-performance liquid chromatography or liquid chromatography–mass spectrometry [19]. Patients were excluded when no [18F]FDG-PET/CT scan was acquired (N = 40) and when patients without a germline mutation were not tested for a somatic mutation (N = 33).
Data acquisition and image reconstruction
PET/CT images were acquired in accordance with the European Association of Nuclear Medicine (EANM) guidelines version 1.0 for tumour PET imaging [20] using a Biograph 40 mCT (Siemens Healthineers). Patients fasted for at least 6 h and serum glucose levels were below 8.0 mmol/L. Image acquisition was started 60 (55–75) minutes after intravenous administration of [18F]FDG. The reconstructed voxel sizes were 3.18 × 3.18 × 3.00 mm3 for PET and ranged from 0.64 × 0.64 × 3 mm3 to 1.27 × 1.27 × 3 mm3 for non-contrast-enhanced low-dose (ld) CT images.
Additional details on data acquisition, image reconstruction, radiomic analysis and the statistical analysis can be found in Online Resource 1: the Image Biomarker Standardisation Initiative (IBSI) Supplementary File 1 [21], which also includes the TRIPOD statement (transparent reporting of a multivariable prediction model for individual prognosis or diagnosis, version October 1, 2020) [22].
Quantitative image analysis
Volume of interest (VOI) delineation
VOI delineation was performed in 3DSlicer version 4.11 (www.slicer.org) [23] and in-house built software implemented in Python 3.7 (Python Software Foundation). Boxing was applied to exclude surrounding [18F]FDG-avid tissues like the kidneys or catecholamine-stimulated brown adipose tissue. Since [18F]FDG uptake of PPGLs can be rather low and heterogeneous, PPGLs were delineated using an isocontour that applies an adaptive threshold of 41% of the SUVpeak, obtained using a sphere of 12 mm diameter [24], corrected for local background (Fig. 1, more details in Supplementary File 1) [25]. This method demonstrated the best agreement between delineated tumour sizes and pathological tumour sizes [26] and allowed the inclusion of most of the vital tumour volume, while minimising the need for boxing. Regions of central necrosis, which were not included by the adaptive threshold algorithm, were not manually added to the VOI, since in PPGL, the classification performance of the radiomic model is not affected by the addition of areas of central necrosis [18]. LdCT images were delineated manually. Lesions were excluded when edges could not be distinguished from intense brown adipose tissue activation (N = 2) [27] or when the minimal size recommendation of 64 voxels per VOI was not met (N = 1) [28].
Image processing
LdCT voxels were interpolated to isotropic voxels (1.5 × 1.5 × 1.5 mm3) using B-spline interpolation, with grids aligned by the input origin. PET images were not interpolated, since the voxels were almost isotropic (3.18 × 3.18 × 3.00 mm3).
Radiomic feature extraction
Radiomic feature extraction was performed in PyRadiomics 3.0 in Python 3.7 (Python Software Foundation) [29]. For both PET and ldCT images, 105 radiomic features were extracted: 18 first-order features, 14 shape features and 73 texture features (Supplementary File 1). In addition, the total lesion glycolysis, the product of the mean SUV and the metabolic tumour volume, was calculated. A fixed bin size of 0.5 g/mL and 25 Hounsfield units was applied for PET and CT images, respectively.
Statistical analysis
Stratified five-fold multinomial logistic regression for the identification of the genetic clusters of PPGLs was performed in R version 3.6.0 (R Foundation for Statistical Computing). Heatmaps were generated using Orange Data Mining version 3.30.2 (University of Ljubljana) [30]. The dataset was split into five equal-sized folds, stratified for the genetic clusters. Each subgroup consecutively served as a test set and the remaining four-fifths of patients served as the training set. Per fold, dimensionality reduction of the radiomic feature set in the training set was performed using redundancy filtering and factor analysis in FMradio (Factor Modelling for Radiomics Data) R-package version 1.1.1 (Fig. 2) [31]. One feature was selected for every ten subjects in the training set [14]. Features were scaled (centred around 0, variance of 1) to avoid that features with the largest scale would dominate the analysis. Redundancy filtering on the Spearman correlation matrix (ρ = 0.95) of the scaled features was performed. Factor analysis was performed on the redundancy filtered correlation matrix using an orthogonal rotation, so that the first factor explained the largest possible variance in the dataset and succeeding factors explained the largest variance in orthogonal directions. The sampling adequacy of the model was determined by the Kaiser-Meier-Olkin measure (KMO, ≥ 0.9) [31]. The factor definitions were determined based on the underlying clusters of features in the different folds. Models were trained for the SUVmax, PET(/CT) factors and imaging variables combined with the biochemical profile. Catecholamines are included as separate dichotomous variables (e.g. adrenergic: yes/no). In addition, the best-performing factor-based model was approximated by a feature-based model using the features underlying the factors, to advance the reproducibility of the radiomic model. The imaging factors and features and the biochemical profile were used as independent variables in multinomial logistic regression. Classification performances were presented as mean multiclass AUCs and mean AUCs between clusters as determined over the five folds for the test sets [32]. A sham experiment was conducted to validate the findings [33]. The outcome labels were randomly shuffled for 100 iterations and mean AUCs were calculated. Randomisation of the outcome labels preserves the distributions and multicollinearity of the radiomic features and the prevalence of the outcome, but it uncouples their potential relation.
Schematic overview of statistical analysis, consisting of stratified 5-fold cross-validation with dimensionality reduction incorporated in the folds. Per fold, scaling was performed (centred around 0, variance of 1), followed by redundancy filtering of the Spearman correlation matrix (ρ = 0.95) and factor analysis using an orthogonal rotation. The factor definitions were determined based on the underlying clusters of features in the different folds
Results
Forty PPGLs in 38 patients were analysed, including thirteen cluster 1, eighteen cluster 2 and nine sporadic PPGLs (Table 1).
The biochemical profile alone could identify the genetic clusters with a mean multiclass AUC of 0.60 (Table 2). The SUVmax alone reached a multiclass AUC of 0.85, with a perfect AUC for distinguishing cluster 1 from cluster 2 PPGLs of 1.00 (Fig. 3). The model with three PET factors showed a slightly improved classification performance with a multiclass AUC of 0.88. The three-factor PET model also showed the highest test AUCs for distinguishing sporadic PPGLs from both cluster 1 and 2 PPGLs (Fig. 3). The (multiclass) AUCs for the model with three PET/CT factors were lower (multiclass AUC: 0.81). The addition of the biochemical profile to the imaging model (SUVmax, three PET factors or three PET/CT factors) increased the (multiclass) AUCs in the training sets but decreased the (multiclass) AUCs in the test sets (Fig. 4). Cluster 1 and 2 PPGLs can be separated with AUCs close to 1.00, cluster 1 and sporadic PPGLs can be distinguished with AUCs around 0.9 and cluster 2 and sporadic PPGLs can be distinguished with AUCs around 0.7. In the sham experiment, no model yielded a (multiclass) AUC different from 0.5 (range: 0.48–0.52).
ROC curves for the SUVmax (green, solid) and PET three-factor model (blue, dashed) between clusters (cluster 1 vs 2, cluster 1 vs sporadic, cluster 2 vs sporadic) as determined by stratified five-fold multinomial logistic regression and the multiclass AUC described by Hand and Till [32]. ROC: receiver operating characteristic, AUC: area under the ROC curve
ROC curves for the biochemical profile alone (grey, solid), PET three-factor model (blue, dashed) and the PET model combined with the biochemical profile (orange, dotted) between clusters (cluster 1 vs 2, cluster 1 vs sporadic, cluster 2 vs sporadic) as determined by stratified five-fold multinomial logistic regression and the multiclass AUC described by Hand and Till [32]. ROC: receiver operating characteristic, AUC: area under the ROC curve
For the PET and PET/CT model, dimensionality reduction retained three factors in every training set (i.e. 32 lesions in all training sets, Table 3). The PET/CT factors corresponded best to the SUVmax, ldCT tumour diameter (3D) and ldCT entropy. The retained factors in the PET model corresponded best to SUVmax, tumour diameter (3D) and grey-level cooccurrence matrix (GLCM) cluster shade. For reproducibility and explainability of the model, these three features were incorporated in a feature-based model, approximating the factor-based model. The feature-based model shows a lower performance than the factor-based model with multiclass AUCs of 0.86 and 0.88 and AUCs for the discrimination of cluster 2 and sporadic PPGLs of 0.63 and 0.72 for the feature-based and factor-based model, respectively (Table 3).
Based on both the PET factors and PET features, cluster 1 PPGLs can be distinguished best from the other clusters (Fig. 5, Fig. 6). Cluster 1 PPGLs show higher means for all features compared to cluster 2 and sporadic PPGLs. Sporadic PPGLs showed imaging characteristics similar to cluster 2 and, to a lesser extent, cluster 1 PPGLs, complicating the differentiation.
Heatmap of the three PET radiomic features (first-order entropy, maximum 3D diameter, GLCM cluster shade) on the y-axis and the PPGLs on the x-axis, clustered by the PPGLs. Cluster 1, cluster 2 and sporadic PPGLs are represented in blue, red and green, respectively. Features were scaled (centred around 0, variance of 1) to avoid that features with the largest scale would dominate the analysis
Discussion
In this study, we assessed the added value of radiomic features derived from PET and non-contrast-enhanced ldCT for the characterisation of the genetic cluster of PPGLs. In our previous research, we showed that the SUVmax alone could already distinguish hereditary cluster 1 and 2 PPGLs with an AUC of 0.91 (95% CI: 0.80–1.00) [11]. This study focused on the identification of both hereditary and somatic cluster 1 and cluster 2 PPGLs, and sporadic PPGLs in a cross-validated radiomic approach. Our findings demonstrate that SUVmax alone already distinguishes cluster 1 and 2 PPGLs with high certainty, but the distinction of clusters 1 and 2 from sporadic PPGLs can be improved moderately by PET radiomics.
Interpretation of the radiomic factors could provide insight into the semantics or tumour phenotype as captured by a PET scan. The first PET factor corresponded to first-order entropy, specifying the randomness in imaging values. Cluster 1 PPGLs show higher mean entropy than cluster 2 and sporadic PPGLs. Entropy is strongly correlated with SUVmax. Genotype-related changes in energy metabolism have been studied using dynamic [18F]FDG-PET/CT scanning [11]. The glucose metabolic rate, phosphorylation rate, vascular blood fraction and SUVmax were all significantly higher in cluster 1 than in cluster 2 and/or sporadic PPGLs. This might be associated with increased expression of hexokinase, which indicates an increase in aerobic glycolysis. The second factor corresponded to the 3D tumour diameter. Cluster 1 PPGLs are typically larger than cluster 2 and, to a lesser extent, sporadic PPGLs. The third factor corresponded to GLCM cluster shade, a feature that measures the skewness of the cooccurrence matrix, thereby characterising the tendency of voxel clusters with similar grey levels. A higher cluster shade implies less clustering and therefore more heterogeneous uptake patterns. Cluster 1 PPGLs show higher cluster shade values than sporadic and cluster 2 PPGLs.
In accordance with the findings of Eisenhofer et al [34], our study showed that the biochemical profile alone could distinguish cluster 1 and 2 PPGLs. However, the biochemical profile could not identify sporadic PPGLs with high certainty. Also, the addition of biochemistry to the imaging models did not improve the classification performance, both showing difficulties differentiating sporadic PPGLs. The training set AUCs were increased, compared to a decrease in the test sets. This indicates overfitting of the models, which can be attributed to the total number of six variables in the combined model, disregarding the criterion of 1 variable per 10 subjects in this small dataset [14]. Also, the addition of ldCT features in dimensionality reduction did not improve the performance of the model. This might indicate that the image quality of the ldCT images was insufficient or the images did not contain characteristics suitable for the differentiation of PPGLs. Differently, the addition of the 105 ldCT features almost doubled the total number of features in the dataset, thereby enlarging the feature space and adding new information, contributing to different factors.
The PET factor model was the best-performing model, but differences with the SUVmax model were small. Therefore, a simplified model with only the SUVmax might be preferred in terms of clinical usability, weighing model performances and the laborious radiomic analysis. Nevertheless, the distinction of sporadic PPGLs from both cluster 1 and 2 PPGLs might be moderately improved by a radiomic model. Cluster 1 and 2 PPGLs generally present distinctive characteristics that can be captured using [18F]FDG-PET imaging. Some of these characteristics can even be assessed visually, like [18F]FDG uptake and tumour size. Sporadic PPGLs, however, appear more heterogeneous, frequently resembling cluster 2 PPGLs and occasionally resembling cluster 1 PPGLs.
Radiomic research in PPGLs on [18F]FDG-PET/CT is limited. Ansquer et al [17] published an article on radiomics in 52 pheochromocytomas with results more or less similar to ours. They also report a higher SUVmax in cluster 1 pheochromocytomas than in cluster 2 and pheochromocytomas without germline mutation. In addition, a model with the features metabolic tumour volume and two texture features could identify germline mutation status with an AUC of 0.95. It is challenging to directly compare these results to ours. Ansquer et al [17] included only pheochromocytomas, which were not tested for somatic mutations, i.e. their sporadic group might have included patients with somatic cluster 1 and 2 mutations, while somatic mutations might have impacted the [18F]FDG uptake as well. In addition, supervised feature selection was performed on the complete dataset, selecting the features with the best association with the outcome measure, and in the final step, 4-fold cross-validation was performed using these selected features. Besides radiomics, proton (1H) nuclear magnetic resonance spectroscopy has been investigated for the identification of genetic clusters of paraganglioma, and the detection of succinate was found to be a highly specific and sensitive hallmark of SDHx mutations [35].
Our study has several strengths and limitations. A strength is that PET/CT images were acquired and reconstructed in accordance with EANM guidelines [20]. The use of these EARL-compliant reconstructions leads to a larger number of reliable, repeatable and reproducible radiomic features [36]. Likewise, radiomic feature extraction was performed and reported conforming to the IBSI recommendations and guidelines, and the TRIPOD statement [21, 22]. Also, unsupervised feature selection, or dimensionality reduction, was performed. In contrast to a supervised approach, dimensionality reduction is not based on the discriminative value of a feature for outcome, but takes into account the interaction of features among themselves and multicollinearity, through which it prevents overfitting of the model [37]. Additionally, dimensionality reduction was incorporated on the training sets per fold instead of on the dataset as a whole, preserving independent test sets. Furthermore, we chose a factor-based over a feature-based approach for the generalisability of our model. Factor analysis was performed incorporated in the folds and instead of selecting features corresponding to the factors, the factors were used as model input. Patterns in corresponding features were compared between folds. In a feature-based approach, insight into these patterns would be limited due to the selection of different features in every fold. In this way, the factor-based approach might advance the generalizability and interpretability of the model and it might provide insight in the semantics or underlying tumour biology of the factors [38]. For the mathematical explainability and reproducibility in the setting of external validation, the PET factor-based model was approximated by a feature-based model. Lastly, we performed a sham experiment to validate our findings [33].
The main limitation of the study is the small sample size. Sample size calculation for radiomics is practically infeasible, as the required number of patients depends on, among other things, the strength of the biological signal, the homogeneity of the data and the complexity of the mathematical model [28]. Orlhac et al [28] state that the minimal sample size is around 70 lesions, in the case of a biological signal that is well reflected by the radiomic features investigated in a cross-validation approach. Our population was smaller than the recommended 70 patients but was unique in terms of patient population. To make the best use of this unique PPGL cohort, we have performed stratified 5-fold cross-validation with strict dimensionality reduction that prevented multicollinearity and retained only one factor for every ten patients in the training set, thereby reducing the overfitting of the model [14].
In conclusion, PET radiomics achieves a better identification of PPGLs compared to biochemistry, SUVmax and/or ldCT radiomics, especially for the differentiation of sporadic PPGLs. However, a model with only SUVmax might be preferred, weighing model performances against the laborious radiomic analysis. Radiomics could only mildly improve the overall clinical classification performance for PPGL, warranting external validation in a larger cohort to validate our findings.
Abbreviations
- [18F]FDG:
-
2-[18F]fluoro-2-deoxy-D-glucose
- AUC:
-
Area under the receiver operating characteristic curve
- CT:
-
Computed tomography
- EANM:
-
European Association of Nuclear Medicine
- GLCM:
-
Grey-level cooccurrence matrix
- IBSI:
-
Image Biomarker Standardisation Initiative
- KMO:
-
Kaiser-Meier-Olkin
- ldCT:
-
Low-dose computed tomography
- MAX:
-
Myc-associated factor X
- NF1:
-
Neurofibromatosis type 1
- PPGL:
-
Pheochromocytoma and paraganglioma
- RET:
-
Rearranged during transfection
- SDHA/B/C/D/AF2:
-
Succinate dehydrogenase subunits A, B, C, D and assembly factor 2
- SUV:
-
Standardised uptake value
- TMEM127:
-
Transmembrane protein 127
- VHL:
-
Von Hippel-Lindau
- VOI:
-
volume of interest
References
Lenders JWM, Eisenhofer G, Mannelli M, Pacak K (2005) Phaeochromocytoma. Lancet 366:665–675
Dahia PLM (2014) Pheochromocytoma and paraganglioma pathogenesis: learning from genetic heterogeneity. Nat Rev Cancer 14:108–119
Favier J, Amar L, Gimenez-Roqueplo AP (2015) Paraganglioma and phaeochromocytoma: from genetics to personalized medicine. Nat Rev Endocrinol 11:101–111
Crona J, Taïeb D, Pacak K (2017) New perspectives on pheochromocytoma and paraganglioma: toward a molecular classification. Endocr Rev 38:489–515
Fishbein L, Leshchiner I, Walter V et al (2017) Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell 31:181–193
Eisenhofer G, Huynh TT, Pacak K et al (2004) Distinct gene expression profiles in norepinephrine- and epinephrine-producing hereditary and sporadic pheochromocytomas: activation of hypoxia-driven angiogenic pathways in von Hippel-Lindau syndrome. Endocr Relat Cancer 11:897–911
Lopez-Jimenez E, Gomez-Lopez G, Leandro-Garcia LJ et al (2010) Research resource: transcriptional profiling reveals different pseudohypoxic signatures in SDHB and VHL-related pheochromocytomas. Mol Endocrinol 24:2382–2391
Lenders JWM, Duh Q-Y, Eisenhofer G et al (2014) Pheochromocytoma and paraganglioma: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab 99:1915–1942
Timmers HJ, Chen CC, Carrasquillo JA et al (2012) Staging and functional characterization of pheochromocytoma and paraganglioma by 18F-fluorodeoxyglucose (18F-FDG) positron emission tomography. J Natl Cancer Inst 104:700–708
van Berkel A, Rao JU, Kusters B et al (2014) Correlation between in vivo 18F-FDG PET and immunohistochemical markers of glucose uptake and metabolism in pheochromocytoma and paraganglioma. J Nucl Med 55:1253–1259
van Berkel A, Vriens D, Visser EP et al (2019) Metabolic subtyping of pheochromocytoma and paraganglioma by (18)F-FDG pharmacokinetics using dynamic PET/CT scanning. J Nucl Med 60:745–751
Limkin EJ, Sun R, Dercle L et al (2017) Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol 28:1191–1206
van Velden FH, Cheebsumon P, Yaqub M et al (2011) Evaluation of a cumulative SUV-volume histogram method for parameterizing heterogeneous intratumoural FDG uptake in non-small cell lung cancer PET studies. Eur J Nucl Med Mol Imaging 38:1636–1647
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Yi X, Guan X, Zhang Y et al (2018) Radiomics improves efficiency for differentiating subclinical pheochromocytoma from lipid-poor adenoma: a predictive, preventive and personalized medical approach in adrenal incidentalomas. EPMA J 9:421–429
Ghosh A, Malla SR, Bhalla AS, Manchanda S, Kandasamy D, Kumar R (2020) Texture analysis of routine T2 weighted fat-saturated images can identify head and neck paragangliomas - a pilot study. Eur J Radiol Open. https://doi.org/10.1016/j.ejro.2020.100248
Ansquer C, Drui D, Mirallié E et al (2020) Usefulness of FDG-PET/CT-based radiomics for the characterization and genetic orientation of pheochromocytomas before surgery. Cancers (Basel). https://doi.org/10.3390/cancers12092424
Noortman WA, Vriens D, Mooij CDY et al (2021) The influence of the exclusion of central necrosis on [18F]FDG PET radiomic analysis. Diagnostics 11:1296
Eisenhofer G, Lattke P, Herberg M et al (2013) Reference intervals for plasma free metanephrines with an age adjustment for normetanephrine for optimized laboratory testing of phaeochromocytoma. Ann Clin Biochem 50:62–69
Boellaard R, O'Doherty MJ, Weber WA et al (2010) FDG PET and PET/CT: EANM procedure guidelines for tumour PET imaging: version 1.0. Eur J Nucl Med Mol Imaging 37:181–200
Zwanenburg A, Leger S, Vallieres M, Lock S (2019) Image biomarker standardisation initiative - feature definitions v11. https://doi.org/10.48550/arXiv.1612.07003
Collins GS, Reitsma JB, Altman DG, Moons KG (2015) Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. https://doi.org/10.1136/bmj.g7594
Fedorov A, Beichel R, Kalpathy-Cramer J et al (2012) 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 30:1323–1341
Wahl RL, Jacene H, Kasamon Y, Lodge MA (2009) From RECIST to PERCIST: evolving considerations for PET response criteria in solid tumors. J Nucl Med 50:122S–150S
Frings V, van Velden FHP, Velasquez LM, Hayes W et al (2014) Repeatability of metabolically active tumor volume measurements with FDG PET/CT in advanced gastrointestinal malignancies: a multicenter study. Radiology 273:539–548
Cheebsumon P, Boellaard R, de Ruysscher D et al (2012) Assessment of tumour size in PET/CT lung cancer studies: PET- and CT-based methods compared to pathology. EJNMMI Res 2:56–56
Steinberg JD, Vogel W, Vegt E (2017) Factors influencing brown fat activation in FDG PET/CT: a retrospective analysis of 15,000+ cases. Br J Radiol. https://doi.org/10.1259/bjr.20170093
Orlhac F, Nioche C, Klyuzhin I, Rahmim A, Buvat I (2021) Radiomics in PET imaging: a practical guide for newcomers. PET Clin 16:597–612
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Demšar J, Curk T, Erjavec A et al (2013) Orange: data mining toolbox in Python. J Mach Learn Res 14:2349–2353
Peeters CF, Übelhör C, Mes SW et al (2019) Stable prediction with radiomics data. https://doi.org/10.48550/arXiv.1903.11696
Hand DJ, Till RJ (2001) A simple generalisation of the area under the ROC curve for multiple class classification problems. Mach Learn 45:171–186
Buvat I, Orlhac F (2019) The dark side of radiomics: on the paramount importance of publishing negative results. J Nucl Med 60:1543–1544
Eisenhofer G, Lenders JW, Timmers H et al (2011) Measurements of plasma methoxytyramine, normetanephrine, and metanephrine as discriminators of different hereditary forms of pheochromocytoma. Clin Chem 57:411–420
Lussey-Lepoutre C, Bellucci A, Morin A et al (2016) In vivo detection of succinate by magnetic resonance spectroscopy as a hallmark of SDHx mutations in paraganglioma. Clin Cancer Res 22:1120–1129
Pfaehler E, van Sluis J, Merema BBJ et al (2020) Experimental multicenter and multivendor evaluation of PET radiomic features performance using 3D printed phantom inserts. J Nucl Med 61:469–476
Nie F, Xu D, Tsang IW, Zhang C (2010) Flexible manifold embedding: a framework for semi-supervised and unsupervised dimension reduction. IEEE Trans Image Process 19:1921–1932
Tomaszewski MR, Gillies RJ (2021) The biological meaning of radiomic features. Radiology 298:505–516
Acknowledgements
The authors want to thank the PET/CT technologists from the Radboud University Medical Center for assistance with the PET/CT scans.
Funding
The dynamic PET study [11] was supported by the European Union Seventh Framework Programme (FP7/2007–2013) under grant agreement 259735 (ENSAT CANCER). No additional funding was received for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Floris H. P. van Velden, Leiden University Medical Center, Leiden, the Netherlands.
Conflicts of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and Biometry
One of the authors has significant statistical expertise.
Ethics approval
This retrospective database study has been reviewed and approved by the Commission on Medical Research Involving Human Subjects Region Arnhem-Nijmegen, the Netherlands (protocol code: 2018-4655, date of approval: 10 December 2018). Informed consent was waived because of the retrospective nature of the study. Patients that objected to the use of their anonymised data were excluded.
Study subjects or cohorts overlap
Some study subjects or cohorts have been previously reported in [10, 11, 18].
Methodology
• Retrospective
• Diagnostic or prognostic study
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Noortman, W.A., Vriens, D., de Geus-Oei, LF. et al. [18F]FDG-PET/CT radiomics for the identification of genetic clusters in pheochromocytomas and paragangliomas. Eur Radiol 32, 7227–7236 (2022). https://doi.org/10.1007/s00330-022-09034-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-09034-5