Abstract
Objectives
To stratify patients with multiple sclerosis (pwMS) based on brain MRI-derived volumetric features using unsupervised machine learning.
Methods
The 3-T brain MRIs of relapsing-remitting pwMS including 3D-T1w and FLAIR-T2w sequences were retrospectively collected, along with Expanded Disability Status Scale (EDSS) scores and long-term (10 ± 2 years) clinical outcomes (EDSS, cognition, and progressive course). From the MRIs, volumes of demyelinating lesions and 116 atlas-defined gray matter regions were automatically segmented and expressed as z-scores referenced to external populations. Following feature selection, baseline MRI-derived biomarkers entered the Subtype and Stage Inference (SuStaIn) algorithm, which estimates subgroups characterized by distinct patterns of biomarker evolution and stages within subgroups. The trained model was then applied to longitudinal MRIs. Stability of subtypes and stage change over time were assessed via Krippendorf’s α and multilevel linear regression models, respectively. The prognostic relevance of SuStaIn classification was assessed with ordinal/logistic regression analyses.
Results
We selected 425 pwMS (35.9 ± 9.9 years; F/M: 301/124), corresponding to 1129 MRI scans, along with healthy controls (N = 148; 35.9 ± 13.0 years; F/M: 77/71) and external pwMS (N = 80; 40.4 ± 11.9 years; F/M: 56/24) as reference populations. Based on 11 biomarkers surviving feature selection, two subtypes were identified, designated as “deep gray matter (DGM)-first” subtype (N = 238) and “cortex-first” subtype (N = 187) according to the atrophy pattern. Subtypes were consistent over time (α = 0.806), with significant annual stage increase (b = 0.20; p < 0.001). EDSS was associated with stage and DGM-first subtype (p ≤ 0.02). Baseline stage predicted long-term disability, transition to progressive course, and cognitive impairment (p ≤ 0.03), with the latter also associated with DGM-first subtype (p = 0.005).
Conclusions
Unsupervised learning modelling of brain MRI-derived volumetric features provides a biologically reliable and prognostically meaningful stratification of pwMS.
Key Points
• The unsupervised modelling of brain MRI-derived volumetric features can provide a single-visit stratification of multiple sclerosis patients.
• The so-obtained classification tends to be consistent over time and captures disease-related brain damage progression, supporting the biological reliability of the model.
• Baseline stratification predicts long-term clinical disability, cognition, and transition to secondary progressive course.
Similar content being viewed by others
Introduction
Brain MRI abnormalities in multiple sclerosis (MS) represent objective indicators of the patient’s biological status, reflecting pathogenetic mechanisms underlying disease evolution [1].
Although a massive body of evidence regarding the biological and clinical relevance of MRI biomarkers has been provided through the years by large-N research studies, their implementation in the single-subject setting and therefore in clinical practice remains challenging [2]. Actually, MRI biomarkers exhibit high variance, resulting from both non-disease-related confounders (e.g., age, sex, other coexisting CNS physiologic and pathologic conditions) and disease-related phenotypic and temporal heterogeneity, thus hampering the definition of absolute cut points and limiting their utility for effective patient stratification.
Over the years, technical advances and the emergence of imaging guidelines [3, 4] have led to the widespread availability of high-quality clinical MRI scans, including sequences with isotropic voxel resolution suitable for volumetric quantifications [5]. Unfortunately, this goldmine of information remains largely unexploited due to the complexity of meaningfully modelling high-dimensional dataset and the frequent lack of associated data reliably reflecting the patients’ clinical status.
Unsupervised machine learning (ML) techniques modelling disease progression based solely on objective biomarker changes, without reliance on a priori clinical information or explicit biomarker thresholds, represent a valuable approach to overcome these issues [6]. Recently, such methods have been applied to primary neurodegenerative disorders of the central nervous system [6,7,8] and showed promising results when translated into the MS scenario with the aim to characterize the disease-specific sequence of clinical and MRI changes [9, 10] or to provide an MRI-driven definition of disease phenotypes [11].
Based on these premises, we applied a recently developed algorithm called Subtype and Staging Inference (SuStaIn), which identifies data-driven subtypes characterized by distinct trajectories of biomarker abnormality accumulation, to clinical MRI scans of a large single-center cohort of relapsing-remitting MS (RRMS) patients. We aimed to demonstrate that, based on a fine-grained volumetric mapping of different brain areas and MS lesions obtained from cross-sectional MRI visits, such approach would provide an accurate patient stratification which is both biologically reliable and prognostically meaningful in the light of longitudinal MRIs and long-term (10-year) motor and cognitive evaluations.
Materials and methods
Participants
In this monocentric retrospective study, brain MRI studies of patients with an MS diagnosis revised according to the 2010 McDonald criteria [12] and a relapsing-remitting (RR) course [13] were screened for eligibility from the radiological and clinical research databases of the MS center of the University of Naples “Federico II,” containing data collected starting from October 2006. Inclusion and exclusion criteria are shown in Fig. 1.
Brain MRI scans of healthy controls (HC) from the same databases and an external population of RRMS patients from the University of Genoa were also selected to develop norms for z-score calculation and select MRI features.
The study was conducted in compliance with the ethical standards and approved by the Ethics Committee “Carlo Romano” of the host institution.
Clinical evaluation
For all patients, clinical disability within 1 week from MRI was estimated using the Expanded Disability Status Scale (EDSS). Patients for whom a long-term clinical and neuropsychological evaluation was available were classified at follow-up (10 ± 2 years from baseline MRI) according to the following: (i) motor disability, ranging from 0 to 3 according to ambulation benchmarks corresponding to EDSS scores < 4.0, ≥ 4.0 and < 6.0, ≥ 6.0 and < 7.0, ≥ 7.0 [14]; (ii) cognitive disability, ranging from 0 to 3 and corresponding to the number of impaired (below 1.5 SD age-, sex- and education-corrected normative values in the healthy population [15]) tests at the Brief International Cognitive Assessment of Multiple Sclerosis (BICAMS) battery [16]; (iii) transition to secondary progressive course [13].
MRI data acquisition and processing
Exams were acquired on the same 3-T scanner (Magnetom Trio, Siemens Healthineers) and included a 3D T1-weighted sequence (≤ 1-mm isotropic voxel size) for volumetric analyses and a T2-weighted FLAIR sequence for the quantification of total demyelinating lesion volume (TLV). Sequence parameters and image processing steps are detailed in the Supplemental Material. Briefly, for all participants, demyelinating lesions were automatically segmented, visually checked, and where needed manually adjusted on FLAIR images to compute TLV, while T1-weighted volumes were used for an atlas-based parcellation of gray matter (GM) into 116 regions defined by the Automated Anatomical Labeling (AAL) atlas [17].
Statistical analysis
A flowchart summarizing data processing and analysis steps is depicted in Fig. 2.
Workflow illustrating the main data processing and analysis steps. Volumes of demyelinating lesions and 116 atlas-defined gray matter regions were automatically segmented based on FLAIR-T2w and T1-w images, respectively. Then, the corresponding volumes were expressed as z-scores with reference to external populations of patients and healthy controls that were also used to select the most altered MRI-derived volumes. Following feature selection, baseline MRI biomarkers entered the Subtype and Stage Inference (SuStaIn) algorithm, using 10-fold cross-validation to determine the optimal number of subtypes and the consistency of progression patterns. Models of up to a maximum of 4 subtypes with z-scores of 1, 2, or 3 for each biomarker were tested (excluding z-score events reached by fewer than 5% of the subjects), corresponding to interpretable levels of mild, moderate, and severe abnormality (color coded from blue to red). The trained model was then fit on all training data and applied to longitudinal MRIs. Finally, the biological reliability and clinical relevance of the SuStaIn classification were assessed in the light of longitudinal MRI scans and clinical outcomes
SuStaIn modelling
SuStaIn is an unsupervised machine learning algorithm combining ideas from clustering and event-based modelling, which describes disease progression as the linear evolution of biomarkers along discrete levels of cumulative alteration, defined in terms of deviation from a reference norm (z-scores) [8]. It simultaneously estimates subgroups characterized by distinct patterns of biomarker evolution and the corresponding trajectories, providing a probabilistic assignment of each subject to a specific subtype and stage within a subtype [8]. Methodological aspects of the SuStaIn algorithm are covered in Young et al [8], while the details of the current analysis are provided in the Supplemental Material.
Briefly, MRI-derived GM and lesion volumes were expressed as z-scores with reference to the HC group and the external RRMS population, respectively, with signs of the z-scores flipped when appropriate so that higher values always represented disease worsening. Baseline MRI scans were used as the training set, while longitudinal visits were reserved for the biological and clinical validation of the initial classification [8].
Only variables associated with a moderate to large (Cohen’s f > 0.25) difference between MS patients and HC were selected and entered the SuStaIn algorithm. Models were evaluated using 10-fold cross-validation (CV) in the training cohort to estimate the optimal number of subtypes and the consistency of the subtype progression patterns: the number of subtypes maximizing the average out-of-sample log-likelihood across CV folds was preferred; the similarity of each subtype progression pattern across CV folds (CVS) was measured using the Bhattacharyya coefficient [8]. Finally, the resulting model was fitted on all subjects of the training cohort and applied to unseen longitudinal MRI scans to assign a probable subtype and stage to each MRI visit.
Testing the biological reliability and clinical relevance of SuStaIn classification
The stability of the SuStaIn subtypes over time was expressed with Krippendorf’s α [18]. To assess the rate of change in disease stage, we fit a multilevel linear regression model in which the SuStaIn stage was the dependent variable and follow-up time (nested within subjects) the predictor, with intercepts and slopes allowed to vary across subjects (random effects). The possible effect of baseline subtype and stage on the slope of longitudinal stage change was assessed by separately adding them (and the corresponding interactions with follow-up time) to the model and testing the significance of interaction terms. Similar models were set up for individual MRI-derived biomarkers.
The clinical relevance of the SuStaIn classification was assessed in relation to both baseline EDSS and long-term clinical outcomes with ordinal/logistic regression (as appropriate) analyses, in which baseline subtype and stage and their interaction, age, and sex were the independent variables. Follow-up time, baseline EDSS, and disease-modifying therapy were included as additional covariates for longitudinal analyses.
Statistical analyses were carried out using the Statistical Package for Social Science (SPSSv25.0, IBM corp.).
Results
Participants
Four hundred and twenty-five RRMS patients (baseline age: 35.9 ± 9.9 years; F/M: 301/124) were selected, corresponding to a total of 1129 MRI visits (2.7 MRI visits per patient, on average; range: 0–9), and a mean follow-up (FU) time of 2.1 years.
MRI scans of 148 HC (age: 35.9 ± 13.0 years; F/M: 77/71) were also selected, along with those of an external population of 80 MS patients (age: 40.4 ± 11.9 years; F/M: 56/24).
Demographic and clinical characteristics of the studied population are reported in Table 1.
Long-term clinical outcomes were available for 178 patients (level of motor disability: 0 = 121, 1 = 35, 2 = 16, 3 = 6; level of cognitive disability: 0 = 81, 1 = 42, 2 = 24, 3 = 31; transition to secondary progressive course: 29 subjects).
SuStaIn model
The volumes of 10 GM regions, including the bilateral anterior cingulate cortices, the right middle cingulate cortex, the bilateral insulae and cunei, the right putamen, and the bilateral thalami, were associated with a moderate to large difference compared with the HC group and were thus fed into the SuStaIn algorithm along with TLV, for a total of 11 biomarkers (Fig. 3, Supplementary Table 1).
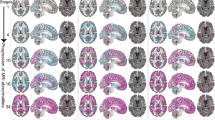
Results of the feature selection procedure. Gray matter regions whose volume survived the feature selection procedure (i.e., associated with a moderate to large effect size at the comparison with healthy controls) are presented, along with a lesion probability map (obtained by summing all the binary lesion masks and dividing by the number of patients, thresholded at 10% probability), all superimposed on axial slices of the average T1w volume in the standard space. Images are in radiological orientation
The two-subtype model yielded the highest average log-likelihood across CV folds (Supplementary Figure 1) and was therefore chosen as the best fitting model for subsequent analyses. When looking at the trajectories of brain damage progression in each subtype, we designated them as follows: (1) the deep gray matter (DGM)-first subtype (56% of subjects, n = 238), characterized by the initial volume loss of subcortical gray matter structures followed by lesion accrual and cortical atrophy; and (2) the cortex-first subtype (44% of subjects, n = 187), characterized by cortical volume loss preceding DGM atrophy and lesion accumulation (Fig. 4).
Positional variance diagrams for the two MRI-driven subtypes. Each entry describes the probability for each biomarker of reaching the color-coded z-score at each SuStaIn stage. The colors represent the degree of abnormality based on the z-score level (blue = mild, z-score of 1; violet = moderate, z-score of 2; red = severe, z-score of 3), while the color shade reflects the uncertainty associated with the corresponding biomarker event. CVS, cross-validation similarity; TLV, total lesion volume
Both progression patterns demonstrated high stability across CV folds, with CVS of 0.91 ± 0.03 and 0.91 ± 0.04 for the DGM-first and cortex-first subtypes, respectively (Supplementary Figure 2).
Patients assigned to the two subtypes had comparable age, sex, and whole brain volume (WBV), while the DGM-first subtype was associated with longer DD (p < 0.001) and higher baseline EDSS score (p = 0.004), SuStaIn stage (p = 0.01), and TLV (p < 0.001) (Table 2).
Biological reliability and clinical relevance
Disease subtypes tended to be consistent over time (Krippendorf’s α = 0.806; CI = 0.752, 0.821), with subtype stability increasing as the probability threshold for the baseline subtype assignment was raised at 95% (177 subjects; α = 0.990; CI = 0.973, 0.998) or 99% (114 subjects; α = 0.990; CI = 0.973, 0.998).
In patients who retained the initial subtype, there was a significant annual increase in disease stage (b = 0.20; SE = 0.05; CI = 0.09, 0.30; p < 0.001), supporting the biological reliability of SuStaIn’s staging, with no significant between-subtype difference (interaction term subtype*follow-up time: b = −0.08; SE = 0.11; CI = −0.29, 0.13; p = 0.48). A significant moderation effect of baseline stage on the relationship between follow-up time and disease stage was observed (interaction term baseline stage*follow-up time: b = −0.05; SE = 0.01; CI = −0.08, −0.02; p = 0.001), corresponding to slopes getting flatter as the baseline stage increased and probably reflecting a plateau effect.
When looking at individual MRI-derived biomarkers, all the GM volumes significantly decreased over time (p ≤ 0.03) (Supplementary Table 2), with significant between-group differences for the left thalamus, corresponding to greater longitudinal atrophy rates in the DGM-first subtype (interaction term subtype*follow-up time: b = 0.05; SE = 0.01; CI = 0.02, 0.07; p = 0.001), and significant plateau effects (the higher the baseline stage, the flatter the slope of longitudinal changes) for the right thalamus (interaction term subtype*follow-up time: b = 0.007; SE = 0.003; CI = 0.002, 0.013; p = 0.006) and the right anterior cingulate cortex (interaction term subtype*follow-up time: b = 0.002; SE = 0.001; CI = 0.001, 0.003; p = 0.002).
As for the relationship with clinical outcomes, baseline EDSS score was positively related with both SuStaIn stage (b = 0.042; p < 0.001) and the DGM-first subtype (b = −0.280; p = 0.02), with baseline stage that also predicted long-term disability (b = 0.030; p = 0.007) and transition to SP course (b = 0.079; p = 0.03). Long-term cognitive impairment was associated with higher baseline stages (b = 0.048; p < 0.001), the DGM-first subtype (b = −0.442; p = 0.005), and their interaction (b = −0.080; p = 0.002) (Table 3).
Discussion
The ambition towards personalized medicine has stimulated increasing efforts to disentangle the inter-subject variability of neurological disorders, integrating information from different biomarkers to identify distinct underlying biological drivers (i.e., biotypes), up to the level of individual patients [19]. In this work, we obtained a biologically consistent and prognostically relevant stratification of RRMS patients based on the unsupervised modeling of brain volumetric features derived from cross-sectional MRI visits.
Using the SuStaIn algorithm, two distinct MRI-driven subtypes were identified, with a latent pattern in which early DGM atrophy and T2 lesion accumulation precede cortical atrophy separated from one in which cortical volume loss precedes DGM atrophy and lesion accrual. These results are essentially in line with the recent work by Eshaghi et al [11], with slight dissimilarities most probably due to the different choices of input features. Indeed, the apparent discrepancy in terms of the number of subtypes is most likely explained by the lack of MRI-derived measures of normal appearing white matter damage in our study, which limited the sensitivity to capture the phenotypic heterogeneity associated with extra-lesional microstructural injury.
On the other hand, the application of a more fine-grained brain parcellation scheme led to a more anatomically precise modelling of GM atrophy, highlighting regions most prominently involved in MS such as the thalami and anterior cingulate, insular, and visual cortices [9]. Interestingly, the fact that distinct disease subtypes remain distinguishable based on the patients’ MRIs even within a relatively clinically homogeneous population confirms the scarce correspondence between clinical and MRI-driven phenotyping, with the latter more closely reflecting disease-related pathogenic mechanisms [11].
Indeed, while patients assigned to the two subtypes did not significantly differ in terms of age, sex, or WBV, the DGM-first subtype was associated with higher DD, stage, and TLV, consistent with the idea of distinct pathogenic mechanisms underpinning cortical and DGM atrophy [20,21,22]. In particular, based on the closer association with TLV, subcortical GM might be more sensitive to the secondary effects of focal demyelination through anterograde/retrograde degeneration, with a prominent role of primary GM neuroinflammation and neurodegeneration in determining cortical atrophy [21,22,23]. Also, the longer DD suggest an earlier diagnosis in patients of the DGM-first subtype, possibly reflecting a shorter prodromal phase [11, 24].
The biological reliability of the MRI-driven classification was further confirmed by the analysis of longitudinal MRI scans, with high subtype stability and significant stage increase over time, reflecting actual temporal progression of brain damage along the estimated paths. Also relevant in terms of biological consistency, moderation analyses suggested plateau effects in the longitudinal trajectories of SuStaIn stage and individual biomarkers (i.e., right thalamus and anterior cingulate cortex atrophy), in line with known temporal patterns of MS-related brain atrophy [25, 26], with steeper thalamic shrinkage rates in the DGM-first subtype.
When assessing the clinical relevance of the SuStaIn classification, higher baseline EDSS scores were independently associated with both higher stages, corresponding to more pronounced brain structural damage, and the DGM-first subtype, a finding consistent with prior evidence pointing at the prominent role of subcortical GM (thalamic, in particular) atrophy in driving disability [27, 28].
As for the prognostic meaning of the MRI-driven stratification, patients in a more advanced position along the damage progression trajectory were more likely to enter the clinically progressive phase in the long term, as well as to suffer greater degrees of motor and cognitive disability, with more severe cognitive impairment also independently associated with the DGM-first subtype. These findings further corroborate the idea that, although cross-sectional in nature, the baseline MRI-driven classification encodes relevant information about future disease evolution, also substantiating the role of subcortical GM atrophy as a relevant anatomical correlate of cognitive disability in MS [29, 30].
Overall, the proposed approach provides insights into MS-related disease mechanisms, confirming and expanding the existing knowledge on MS physiopathology. But even more interestingly, it condenses this complex information at the patient level in simple and intuitive measures which are easily obtainable from single-visit conventional MRI scans and correlate with clinical measures of disease severity and progression. Contextualizing the information contained in individual brain MRIs in the frame of disease patterns estimated in a reference population of MS patients, such stratification holds potential for effectively linking MS research to the single-subject setting, with relevant implications for both clinical trials and routine practice.
Our work is not without limitations. While the monocentric nature of the study reduces the data heterogeneity related to scanner/center effects, it also limits the model generalizability, prompting larger studies on multicentric datasets. Furthermore, increasing the sample size would also allow for a higher dimensional (and more accurate) representation of MS pathology, possibly including additional biomarkers from spinal cord imaging or from other advanced MRI techniques encoding relevant information about the brain microstructure (e.g., diffusion MRI, quantitative MRI) [20, 31] or function (e.g., functional MRI) [32].
In conclusion, through the unsupervised modelling of volumetric features derived from brain MRI scans, we obtained a biologically reliable and prognostically meaningful single-visit classification of MS patients, potentially offering a powerful tool for subjects’ stratification in both trial design and clinical practice.
Change history
22 July 2022
In this article the following funding information was added: Open access funding provided by Università degli Studi di Napoli Federico II within the CRUI-CARE Agreement.
Abbreviations
- DD:
-
Disease duration
- DGM:
-
Deep gray matter
- DMT:
-
Disease-modifying therapy
- EDSS:
-
Expanded Disability Status Scale
- GM:
-
Gray matter
- HC:
-
Healthy controls
- MS:
-
Multiple sclerosis
- RR:
-
Relapsing remitting
- TLV:
-
Total lesion volume
- WBV:
-
Whole brain volume
References
Filippi M, Brück W, Chard D et al (2019) Association between pathological and MRI findings in multiple sclerosis. Lancet Neurol 18:198–210
Vermersch P, Berger T, Gold R et al (2016) The clinical perspective: how to personalise treatment in MS and how may biomarkers including imaging contribute to this? Mult Scler 22:18–33
Traboulsee A, Simon JH, Stone L et al (2016) Revised recommendations of the Consortium of MS Centers Task Force for a standardized mri protocol and clinical guidelines for the diagnosis and follow-up of multiple sclerosis. AJNR Am J Neuroradiol 37:394–401
Wattjes MP, Rovira A, Miller D et al (2015) Evidence-based guidelines: MAGNIMS consensus guidelines on the use of MRI in multiple sclerosis--establishing disease prognosis and monitoring patients. Nat Rev Neurol 11:597–606
Pontillo G, Cocozza S, Di Stasi M et al (2020) 2D linear measures of ventricular enlargement may be relevant markers of brain atrophy and long-term disability progression in multiple sclerosis. Eur Radiol 30:3813–3822
Young AL, Oxtoby NP, Daga P et al (2014) A data-driven model of biomarker changes in sporadic Alzheimer's disease. Brain 137:2564–2577
Fonteijn HM, Modat M, Clarkson MJ et al (2012) An event-based model for disease progression and its application in familial Alzheimer's disease and Huntington's disease. Neuroimage 60:1880–1889
Young AL, Marinescu RV, Oxtoby NP et al (2018) Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with Subtype and Stage Inference. Nat Commun 9:4273
Eshaghi A, Marinescu RV, Young AL et al (2018) Progression of regional grey matter atrophy in multiple sclerosis. Brain 141:1665–1677
Dekker I, Schoonheim MM, Venkatraghavan V et al (2021) The sequence of structural, functional and cognitive changes in multiple sclerosis. Neuroimage Clin 29:102550
Eshaghi A, Young AL, Wijeratne PA et al (2021) Identifying multiple sclerosis subtypes using unsupervised machine learning and MRI data. Nat Commun 12:2078
Polman CH, Reingold SC, Banwell B et al (2011) Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol 69:292–302
Lublin FD, Reingold SC, Cohen JA et al (2014) Defining the clinical course of multiple sclerosis: the 2013 revisions. Neurology 83:278–286
Confavreux C, Vukusic S (2006) Age at disability milestones in multiple sclerosis. Brain 129:595–605
Goretti B, Niccolai C, Hakiki B et al (2014) The Brief International Cognitive Assessment for Multiple Sclerosis (BICAMS): normative values with gender, age and education corrections in the Italian population. BMC Neurol 14:171
Benedict RH, Amato MP, Boringa J et al (2012) Brief International Cognitive Assessment for MS (BICAMS): international standards for validation. BMC Neurol 12:55
Tzourio-Mazoyer N, Landeau B, Papathanassiou D et al (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273–289
Hayes AF, Krippendorff K (2007) Answering the call for a standard reliability measure for coding data. Communication Methods and Measures 1:77–89
Gibbs RM, Lipnick S, Bateman JW et al (2018) Toward precision medicine for neurological and neuropsychiatric disorders. Cell Stem Cell 23:21–24
Pontillo G, Cocozza S, Lanzillo R et al (2019) Determinants of deep gray matter atrophy in multiple sclerosis: a multimodal MRI study. AJNR Am J Neuroradiol 40:99–106
Calabrese M, Magliozzi R, Ciccarelli O, Geurts JJG, Reynolds R, Martin R (2015) Exploring the origins of grey matter damage in multiple sclerosis. Nature Reviews Neuroscience 16:147–158
Geurts JJ, Barkhof F (2008) Grey matter pathology in multiple sclerosis. Lancet Neurol 7:841–851
Ruggieri S, Petracca M, Miller A et al (2015) Association of deep gray matter damage with cortical and spinal cord degeneration in primary progressive multiple sclerosis. JAMA Neurol 72:1466–1474
Wijnands JMA, Kingwell E, Zhu F et al (2017) Health-care use before a first demyelinating event suggestive of a multiple sclerosis prodrome: a matched cohort study. Lancet Neurol 16:445–451
Pagani E, Rocca MA, Gallo A et al (2005) Regional brain atrophy evolves differently in patients with multiple sclerosis according to clinical phenotype. AJNR Am J Neuroradiol 26:341–346
Fisher E, Lee JC, Nakamura K, Rudick RA (2008) Gray matter atrophy in multiple sclerosis: a longitudinal study. Ann Neurol 64:255–265
Eshaghi A, Prados F, Brownlee WJ et al (2018) Deep gray matter volume loss drives disability worsening in multiple sclerosis. Ann Neurol 83:210–222
Minagar A, Barnett MH, Benedict RH et al (2013) The thalamus and multiple sclerosis: modern views on pathologic, imaging, and clinical aspects. Neurology 80:210–219
Houtchens MK, Benedict RH, Killiany R et al (2007) Thalamic atrophy and cognition in multiple sclerosis. Neurology 69:1213–1223
Petracca M, Pontillo G, Moccia M et al (2021) Neuroimaging correlates of cognitive dysfunction in adults with multiple sclerosis. Brain Sci 11:346
Pontillo G, Petracca M, Monti S et al (2021) Unraveling deep gray matter atrophy and iron and myelin changes in multiple sclerosis. AJNR Am J Neuroradiol. https://doi.org/10.3174/ajnr.A7093
Cocozza S, Pontillo G, Russo C et al (2018) Cerebellum and cognition in progressive MS patients: functional changes beyond atrophy? J Neurol 265:2260–2266
Funding
Open access funding provided by Università degli Studi di Napoli Federico II within the CRUI-CARE Agreement. The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Giuseppe Pontillo, MD.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• observational
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 711 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pontillo, G., Penna, S., Cocozza, S. et al. Stratification of multiple sclerosis patients using unsupervised machine learning: a single-visit MRI-driven approach. Eur Radiol 32, 5382–5391 (2022). https://doi.org/10.1007/s00330-022-08610-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08610-z