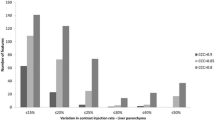
Abstract
Objectives
Following the craze for radiomic features (RF), their lack of reliability raised the question of the generalizability of classification models. Inter-site harmonization of images therefore becomes a central issue. We compared RF harmonization processing designed to detect liver diseases in CT images.
Methods
We retrospectively analyzed 76 multi-center portal CT series of non-diseased (NDL) and diseased liver (DL) patients. In each series, we positioned volumes of interest in spleen and liver, then extracted 9 RF (histogram and texture). We evaluated two RF harmonization approaches. First, in each series, we computed the Z-score of liver measurements based on those computed in the spleen. Second, we evaluated the ComBat method according to each imaging center; parameters were computed in the spleen and applied to the liver. We compared RF distributions and classification performances before/after harmonization. We classified NDL versus spleen and versus DL tissues.
Results
The RF distributions were all different between liver and spleen (p < 0.05). The Z-score harmonization outperformed for the detection of liver versus spleen: AUC = 93.1% (p < 0.001). For the detection of DL versus NDL, in a case/control setting, we found no differences between the harmonizations: mean AUC = 73.6% (p = 0.49). Using the whole datasets, the performances were improved using ComBat (p = 0.05) AUC = 82.4% and degraded with Z-score AUC = 67.4% (p = 0.008).
Conclusions
Data harmonization requires to first focus on data structuring to not degrade the performances of subsequent classifications. Liver tissue classification after harmonization of spleen-based RF is a promising strategy for improving the detection of DL tissue.
Key Points
• Variability of acquisition parameter makes radiomics of CT features non-reproducible.
• Data harmonization can help circumvent the inter-site variability of acquisition protocols.
• Inter-site harmonization must be carefully implemented and requires designing consistent data sets.





Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the curve
- CI:
-
Confidence interval
- CT:
-
Computed tomography
- FOV:
-
Field of view
- GLCM:
-
Gray-level co-occurrence matrix
- GLZLM:
-
Gray-level zone length matrix
- HCC:
-
Hepatocellular carcinoma
- HU:
-
Hounsfield unit
- ICC:
-
Intrahepatic cholangiocarcinoma
- KNN:
-
K-nearest neighbor
- LDA:
-
Linear discriminant analysis
- NGLDM:
-
Neighboring gray-level dependence matrix
- Nnet:
-
Neural network
- QIB:
-
Quantitative imaging biomarker
- RF:
-
Random forest
- ROC:
-
Receiver operating characteristic
- ROI:
-
Region of interest
- SVM:
-
Support vector machine
- VOI:
-
Volume of interest
References
Alic L, Niessen WJ, Veenland JF (2014) Quantification of heterogeneity as a biomarker in tumor imaging: a systematic review. PLoS One 9:e110300. https://doi.org/10.1371/journal.pone.0110300
Traverso A, Wee L, Dekker A, Gillies R (2018) Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys 102:1143–1158. https://doi.org/10.1016/j.ijrobp.2018.05.053
Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ (2017) CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics 37:1483–1503. https://doi.org/10.1148/rg.2017170056
Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D (2017) Characterization of PET/CT images using texture analysis: the past, the present… any future? Eur J Nucl Med Mol Imaging 44:151–165. https://doi.org/10.1007/s00259-016-3427-0
Eisenhauer EA, Therasse P, Bogaerts J et al (2009) New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45:228–247. https://doi.org/10.1016/j.ejca.2008.10.026
Kumar V, Gu Y, Basu S et al (2012) Radiomics: the process and the challenges. Magn Reson Imaging 30:1234–1248. https://doi.org/10.1016/j.mri.2012.06.010
O’Connor JPB, Aboagye EO, Adams JE et al (2017) Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 14:169–186. https://doi.org/10.1038/nrclinonc.2016.162
Kalendralis P, Traverso A, Shi Z et al (2019) Multicenter CT phantoms public dataset for radiomics reproducibility tests. Med Phys 46:1512–1518. https://doi.org/10.1002/mp.13385
Mackin D, Fave X, Zhang L et al (2015) Measuring CT scanner variability of radiomics features. Invest Radiol 50:757–765. https://doi.org/10.1097/RLI.0000000000000180
Simon P, Uma V (2018) Review of texture descriptors for texture classification. Data Eng Intell Comp 542:159–176. https://doi.org/10.1007/978-981-10-3223-3_15
Ger RB, Zhou S, Chi PCM et al (2018) Comprehensive investigation on controlling for CT imaging variabilities in radiomics studies. Sci Rep 8:1–14. https://doi.org/10.1038/s41598-018-31509-z
Berenguer R, Del Rosario Pastor-Juan M, Canales-Vázquez J et al (2018) Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology 288:407–415. https://doi.org/10.1148/radiol.2018172361
Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127. https://doi.org/10.1093/biostatistics/kxj037
Orlhac F, Frouin F, Nioche C, Ayache N, Buvat I (2019) Validation of a method to compensate multicenter effects affecting CT radiomics. Radiology 291:53–59. https://doi.org/10.1148/radiol.2019182023
Nioche C, Orlhac F, Boughdad S et al (2018) Lifex: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res 78:4786–4789. https://doi.org/10.1158/0008-5472.CAN-18-0125
Orlhac F, Soussan M, Chouahnia K, Martinod E, Buvat I (2015) 18F-FDG PET-derived textural indices reflect tissue-specific uptake pattern in non-small cell lung cancer. PLoS One 10:1–16. https://doi.org/10.1371/journal.pone.0145063
Caramella C, Allorant A, Orlhac F et al (2018) Can we trust the calculation of texture indices of CT images? A phantom study. Med Phys 45:1529–1536. https://doi.org/10.1002/mp.12809
Burke Ó, Benton S, Szafranski P et al (2016) Extending the scope of pooled analyses of individual patient biomarker data from heterogeneous laboratory platforms and cohorts using merging algorithms. Pregnancy Hypertens 6:53–59. https://doi.org/10.1016/j.preghy.2015.12.002
Mergo PJ, Ros PR, Buetow PC, Buck JL (1994) Diffuse disease of the liver: radiologic-pathologic correlation. Radiographics 14:1291–1307. https://doi.org/10.1148/radiographics.14.6.7855342
Ahmed AM, Ebid ME, Ajlan AM, Al-Mallah MH (2017) Low-dose attenuation correction in diagnosis of non-alcoholic fatty liver disease. Abdom Radiol (NY) 42:2454–2459. https://doi.org/10.1007/s00261-017-1166-8
Team RDC (2011) R: a language and environment for statistical computing. R Found Stat Comput 1:409
Nygaard V, Rødland EA, Hovig E (2016) Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses. Biostatistics 17:29–39. https://doi.org/10.1093/biostatistics/kxv027
Perrin T, Midya A, Yamashita R et al (2018) Short-term reproducibility of radiomic features in liver parenchyma and liver malignancies on contrast-enhanced CT imaging. Abdom Radiol (NY) 43:3271–3278. https://doi.org/10.1007/s00261-018-1600-6
Brenet Defour L, Mulé S, Tenenhaus A et al (2019) Hepatocellular carcinoma: CT texture analysis as a predictor of survival after surgical resection. Eur Radiol 29:1231–1239. https://doi.org/10.1007/s00330-018-5679-5
Chen S, Zhu Y, Liu Z, Liang C (2017) Texture analysis of baseline multiphasic hepatic computed tomography images for the prognosis of single hepatocellular carcinoma after hepatectomy: A retrospective pilot study. Eur J Radiol 90:198–204. https://doi.org/10.1016/j.ejrad.2017.02.035
Park HJ, Kim JH, Choi S et al (2017) Prediction of therapeutic response of hepatocellular carcinoma to transcatheter arterial chemoembolization based on pretherapeutic dynamic CT and textural findings. AJR Am J Roentgenol 209:W211–W220. https://doi.org/10.2214/AJR.16.17398
Chang C-C, Chen H-H, Chang Y-C et al (2017) Computer-aided diagnosis of liver tumors on computed tomography images. Comput Methods Programs Biomed 145:45–51. https://doi.org/10.1016/j.cmpb.2017.04.008
Reinert CP, Kloth C, Fritz J, Nikolaoua K, Horgera M (2018) Discriminatory CT-textural features in splenic infiltration of lymphoma versus splenomegaly in liver cirrhosis versus normal spleens in controls and evaluation of their role for longitudinal lymphoma monitoring. Eur J Radiol 104:129–135. https://doi.org/10.1016/j.ejrad.2018.05.010
Cozzi L, Dinapoli N, Fogliata A et al (2017) Radiomics based analysis to predict local control and survival in hepatocellular carcinoma patients treated with volumetric modulated arc therapy. BMC Cancer 17:1–10. https://doi.org/10.1186/s12885-017-3847-7
Beckers RCJ, Trebeschi S, Maas M et al (2018) CT texture analysis in colorectal liver metastases and the surrounding liver parenchyma and its potential as an imaging biomarker of disease aggressiveness, response and survival. Eur J Radiol 102:15–21. https://doi.org/10.1016/j.ejrad.2018.02.031
Lee SJ, Zea R, Kim DH, Lubner MG, Deming DA, Pickhardt PJ (2018) CT texture features of liver parenchyma for predicting development of metastatic disease and overall survival in patients with colorectal cancer. Eur Radiol 28:1520–1528. https://doi.org/10.1007/s00330-017-5111-6
Acknowledgments
We gratefully acknowledge Sebastien Patriti, Mahaut Macrez, and Nicholas Terry for their support in this study.
Funding
This study did not receive any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Hubert Beaumont.
Conflict of interest
The authors of this manuscript, Hubert Beaumont, as employee, declares relationships with Median Technologies.
Other authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was obtained from patients.
Ethical approval
For our retrospective study that did not include interaction or intervention with human subjects or include any access to identifiable private information, no IRB approval was required.
Study subjects or cohorts overlap
Study subjects or cohort were involved in other studies
Methodology
• Retrospective
• Experimental
• Performed at multiple institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 574 kb)
Rights and permissions
About this article
Cite this article
Beaumont, H., Iannessi, A., Bertrand, AS. et al. Harmonization of radiomic feature distributions: impact on classification of hepatic tissue in CT imaging. Eur Radiol 31, 6059–6068 (2021). https://doi.org/10.1007/s00330-020-07641-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07641-8