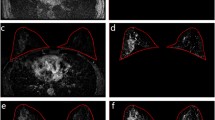
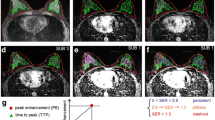
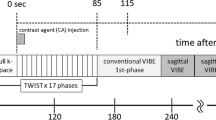
Abstract
Purpose
To assess the diagnostic performance of radiomic analysis using high temporal resolution (HTR)-dynamic contrast enhancement (DCE) MR sequences compared to BI-RADS analysis to distinguish benign from malignant breast lesions.
Materials and methods
We retrospectively analyzed data from consecutive women who underwent breast MRI including HTR-DCE MR sequencing for abnormal enhancing lesions and who had subsequent pathological analysis at our tertiary center. Semi-quantitative enhancement parameters and textural features were extracted. Temporal change across each phase of textural features in HTR-DCE MR sequences was calculated and called “kinetic textural parameters.” Statistical analysis by LASSO logistic regression and cross validation was performed to build a model. The diagnostic performance of the radiomic model was compared to the results of BI-RADS MR score analysis.
Results
We included 117 women with a mean age of 54 years (28–88). Of the 174 lesions analyzed, 75 were benign and 99 malignant. Seven semi-quantitative enhancement parameters and 57 textural features were extracted. Regression analysis selected 15 significant variables in a radiomic model (called “malignant probability score”) which displayed an AUC = 0.876 (sensitivity = 0.98, specificity = 0.52, accuracy = 0.78). The performance of the malignant probability score to distinguish benign from malignant breast lesions (AUC = 0.876, 95%CI 0.825–0.925) was significantly better than that of BI-RADS analysis (AUC = 0.831, 95%CI 0.769–0.892). The radiomic model significantly reduced false positives (42%) with the same number of missed cancers (n = 2).
Conclusion
A radiomic model including kinetic textural features extracted from an HTR-DCE MR sequence improves diagnostic performance over BI-RADS analysis.
Key Points
• Radiomic analysis using HTR-DCE is of better diagnostic performance (AUC = 0.876) than conventional breast MRI reading with BI-RADS (AUC = 0.831) (p < 0.001).
• A radiomic malignant probability score under 19.5% gives a negative predictive value of 100% while a malignant probability score over 81% gives a positive predictive value of 100%.
• Kinetic textural features extracted from HTR-DCE-MRI have a major role to play in distinguishing benign from malignant breast lesions.






Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the receiver operating characteristic curve
- BI-RADS:
-
Breast Imaging Reporting and Data System
- CCC:
-
Concordance correlation coefficient
- CEROG:
-
Comité d’Ethique de la Recherche en Obstetrique et Gynecologie
- DCE:
-
Dynamic contrast enhancement
- DCIS:
-
Ductal carcinomas in situ
- DISCO:
-
Differential Subsampling with Cartesian Ordering
- EA:
-
Enhancement amplitude
- EI:
-
Enhancement integral
- GLCM:
-
Gray-level co-occurrence matrix
- GLDM:
-
Gray-level dependence matrix
- GLRLM:
-
Gray-level run length matrix
- GLSZM:
-
Gray-level size zone matrix
- HTR:
-
High temporal resolution
- ICC:
-
Intra-class correlation coefficient
- IDC:
-
Invasive ductal carcinoma
- Imc1:
-
Informational measure of correlation 1
- LASSO:
-
Least Absolute Shrinkage and Selection Operator
- MR:
-
Magnetic resonance
- MSI:
-
Maximum slope of increase
- PACS:
-
Picture archiving and communication system
- RLNN:
-
Run length non-uniformity normalized
- Rmax:
-
Maximum of enhancement
- RmaxTiming:
-
Timing of maximum of enhancement
- ROC:
-
Receiver operating characteristic
- ROI:
-
Region of interest
- SPGR:
-
Spoiled gradient recalled
- STD:
-
Standard deviation of signal intensity
- THR:
-
Time of half rising
- WIR:
-
Wash-in-rate
References
Sardanelli F, Boetes C, Borisch B et al (2010) Magnetic resonance imaging of the breast: recommendations from the EUSOMA working group. Eur J Cancer 1990 46(8):1296–1316
Mann RM, Cho N, Moy L (2019) Breast MRI: state of the art. Radiology. 292(3):520–536
Kuhl C (2007) The current status of breast MR imaging. Part I. Choice of technique, image interpretation, diagnostic accuracy, and transfer to clinical practice. Radiology. 244(2):356–378
Mann RM, Mus RD, van Zelst J, Geppert C, Karssemeijer N, Platel B (2014) A novel approach to contrast-enhanced breast magnetic resonance imaging for screening: high-resolution ultrafast dynamic imaging. Invest Radiol 49(9):579–585
Milon A, Vande Perre S, Poujol J et al (2019) Abbreviated breast MRI combining FAST protocol and high temporal resolution (HTR) dynamic contrast enhanced (DCE) sequence. Eur J Radiol 117:199–208
Parekh VS, Jacobs MA (2017) Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer 3:43
Rizzo S, Botta F, Raimondi S et al (2018) Radiomics of high-grade serous ovarian cancer: association between quantitative CT features, residual tumour and disease progression within 12 months. Eur Radiol 28(11):4849–4859
Wu G, Woodruff HC, Sanduleanu S et al (2020) Preoperative CT-based radiomics combined with intraoperative frozen section is predictive of invasive adenocarcinoma in pulmonary nodules: a multicenter study. Eur Radiol 30(5):2680–2691
Thomassin-Naggara I, Soualhi N, Balvay D, Darai E, Cuenod C-A (2017) Quantifying tumor vascular heterogeneity with DCE-MRI in complex adnexal masses: a preliminary study. J Magn Reason Imaging. https://doi.org/10.1002/jmri.25707
Chang Y-C, Huang C-S, Liu Y-J, Chen J-H, Lu Y-S, Tseng W-YI (2004) Angiogenic response of locally advanced breast cancer to neoadjuvant chemotherapy evaluated with parametric histogram from dynamic contrast-enhanced MRI. Phys Med Biol 49(16):3593–3602
Ahmed A, Gibbs P, Pickles M, Turnbull L (2013) Texture analysis in assessment and prediction of chemotherapy response in breast cancer. J Magn Reason Imaging 38(1):89–101
Ashraf A, Gaonkar B, Mies C et al (2015) Breast DCE-MRI kinetic heterogeneity tumor markers: preliminary associations with neoadjuvant chemotherapy response. Transl Oncol 8(3):154–162
Bhooshan N, Giger ML, Jansen SA, Li H, Lan L, Newstead GM (2010) Cancerous breast lesions on dynamic contrast-enhanced MR images: computerized characterization for image-based prognostic markers. Radiology. 254(3):680–690
Kim J-H, Ko ES, Lim Y et al (2017) Breast cancer heterogeneity: MR imaging texture analysis and survival outcomes. Radiology. 282(3):665–675
Parikh J, Selmi M, Charles-Edwards G et al (2014) Changes in primary breast cancer heterogeneity may augment midtreatment MR imaging assessment of response to neoadjuvant chemotherapy. Radiology. 272(1):100–112
Thibault G, Tudorica A, Afzal A et al (2017) DCE-MRI texture features for early prediction of breast cancer therapy response. Tomography 3(1):23–32
Wu J, Cao G, Sun X et al (2018) Intratumoral spatial heterogeneity at perfusion MR imaging predicts recurrence-free survival in locally advanced breast cancer treated with neoadjuvant chemotherapy. Radiology. 288(1):26–35
Wu J, Gong G, Cui Y, Li R (2016) Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J Magn Reason Imaging 44(5):1107–1115
Braman NM, Etesami M, Prasanna P et al (2017) Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res 19(1):57
Fan M, Li H, Wang S, Zheng B, Zhang J, Li L (2017) Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLoS One 12(2):e0171683
Fan M, Cheng H, Zhang P et al (2018) DCE-MRI texture analysis with tumor subregion partitioning for predicting Ki-67 status of estrogen receptor-positive breast cancers. J Magn Reason Imaging 48(1):237–247
Li H, Zhu Y, Burnside ES et al (2016) MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology. 281(2):382–391
Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C (2019) Radiomic versus convolutional neural networks analysis for classification of contrast-enhancing lesions at multiparametric breast MRI. Radiology. 290(2):290–297
Fan M, Zhang P, Wang Y et al (2019) Radiomic analysis of imaging heterogeneity in tumours and the surrounding parenchyma based on unsupervised decomposition of DCE-MRI for predicting molecular subtypes of breast cancer. Eur Radiol 29(8):4456–4467
Lo Gullo R, Daimiel I, Rossi Saccarelli C et al (2020) Improved characterization of sub-centimeter enhancing breast masses on MRI with radiomics and machine learning in BRCA mutation carriers. Eur Radiol. https://doi.org/10.1007/s00330-020-06991-7
D’Amico NC, Grossi E, Valbusa G et al (2020) A machine learning approach for differentiating malignant from benign enhancing foci on breast MRI. Eur Radiol Exp 4(1):5
Saranathan M, Rettmann DW, Hargreaves BA, Clarke SE, Vasanawala SS (2012) DIfferential Subsampling with Cartesian Ordering (DISCO): a high spatio-temporal resolution Dixon imaging sequence for multiphasic contrast enhanced abdominal imaging. J Magn Reson Imaging 35(6):1484–1492
Morris EA, Comstock CE, Lee CH (2013) ACR BI-RADS Atlas - Breast Imaging Reporting and Data System Atlas. American College of Radiology, Reston, VA
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 44(3):837–845
Thomassin-Naggara I, Bazot M, Daraï E, Callard P, Thomassin J, Cuenod CA (2008) Epithelial ovarian tumors: value of dynamic contrast-enhanced MR imaging and correlation with tumor angiogenesis. Radiology. 248(1):148–159
Thomassin-Naggara I, Daraï E, Cuenod CA, Rouzier R, Callard P, Bazot M (2008) Dynamic contrast-enhanced magnetic resonance imaging: a useful tool for characterizing ovarian epithelial tumors. J Magn Reson Imaging 28(1):111–120
Yushkevich PA, Piven J, Hazlett HC et al (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3):1116–1128
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77(21):e104–e107
Zwanenburg A, Leger S, Vallières M, Löck S, Initiative for the IBS (2016) Image biomarker standardisation initiative. ArXiv161207003 Cs. [cited 2018 Mar 25]; Available from: http://arxiv.org/abs/1612.07003. Accessed 25 March 2018
Duron L, Balvay D, Vande Perre S et al (2019) Gray-level discretization impacts reproducible MRI radiomics texture features. PLoS One 14(3):e0213459
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Internet]. 2013. Available from: http://www.R-project.org/. Accessed 25 Feb 2019
Tibshirani R (1996) Regression shrinkage and selection via the Lasso. J R Stat Soc Ser B Methodol 58(1):267–288
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14(12):749–762
Liang C, Cheng Z, Huang Y et al (2018) An MRI-based radiomics classifier for preoperative prediction of Ki-67 status in breast cancer. Acad Radiol 25(9):1111–1117
Harvey SC, Di Carlo PA, Lee B, Obadina E, Sippo D, Mullen L (2016) An abbreviated protocol for high-risk screening breast MRI saves time and resources. J Am Coll Radiol 13(11S):R74–R80
Kuhl CK (2018) Abbreviated breast MRI for screening women with dense breast: the EA1141 trial. Br J Radiol 91(1090):20170441
Cuenod CA, Balvay D (2013) Perfusion and vascular permeability: basic concepts and measurement in DCE-CT and DCE-MRI. Diagn Interv Imaging 94(12):1187–1204
Pradel C, Siauve N, Bruneteau G et al (2003) Reduced capillary perfusion and permeability in human tumour xenografts treated with the VEGF signalling inhibitor ZD4190: an in vivo assessment using dynamic MR imaging and macromolecular contrast media. Magn Reson Imaging 21(8):845–851
Folkman J. Tumor angiogenesis. In: Klein G, Weinhouse S, editors. Advances in cancer research. Academic Press; 1985 [cited 2018 Jun 6]. p. 175–203. Available from: http://www.sciencedirect.com/science/article/pii/S0065230X0860946X. Accessed 06 June 2018
McDonald DM, Choyke PL (2003) Imaging of angiogenesis: from microscope to clinic. Nat Med 9(6):713–725
Fukumura D, Jain RK (2008) Imaging angiogenesis and the microenvironment. APMIS 116(7–8):695–715
Bergers G, Benjamin LE (2003) Tumorigenesis and the angiogenic switch. Nat Rev Cancer 3(6):401–410
Reig B, Heacock L, Geras KJ, Moy L (2019) Machine learning in breast MRI. J Magn Reason Imaging. https://doi.org/10.1002/jmri.26852
Agner SC, Soman S, Libfeld E et al (2011) Textural kinetics: a novel dynamic contrast-enhanced (DCE)-MRI feature for breast lesion classification. J Digit Imaging 24(3):446–463
Milenković J, Dalmış MU, Žgajnar J, Platel B (2017) Textural analysis of early-phase spatiotemporal changes in contrast enhancement of breast lesions imaged with an ultrafast DCE-MRI protocol. Med Phys 44(9):4652–4664
Zhou X, Gao F, Duan S et al (2020) Radiomic features of Pk-DCE MRI parameters based on the extensive Tofts model in application of breast cancer. Phys Eng Sci Med 43(2):517–524
Valdora F, Houssami N, Rossi F, Calabrese M, Tagliafico AS (2018) Rapid review: radiomics and breast cancer. Breast Cancer Res Treat 169(2):217–229
Whitney HM, Li H, Ji Y, Liu P, Giger ML (2020) Harmonization of radiomic features of breast lesions across international DCE-MRI datasets. J Med Imaging (Bellingham) 7(1):012707
Rotili A, Trimboli RM, Penco S et al (2020) Double reading of diffusion-weighted magnetic resonance imaging for breast cancer detection. Breast Cancer Res Treat 180(1):111–120
Acknowledgments
Nicolas Mion Eng. and Julie Poujol, PhD.
Funding
Saskia Vande Perre has received a Young Award from the Société Francaise de Radiologie.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Isabelle Thomassin-Naggara.
Conflict of interest
Saskia Vande Perre, Loic Duron, Audrey Milon, Asma Bekhouche, Daniel Balvay, and François H. Cornelis: no relationships with any companies, whose products or services may be related to the subject matter of the article.
Laure Fournier: Receipt of grants/research supports (related with the subject matter of the article) with Philips, ArianaPharma, Evolucare, Invectys. Receipt of honoraria or consultation fees (not related with the subject) with Novartis, Janssen, and Sanofi. Speaker fees (not related with the subject) with Novartis, Bayer, Janssen, Sanofi, Pfizer, GE Healthcare.
Isabelle Thomassin - Naggara: Remunerated lecture with GE, Hologic, Canon, Guerbet, and one participation to board expert meeting (Siemens).
Statistics and biometry
Nicolas Mion, engineer, kindly provided statistical advice for this manuscript.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
CEROG 2018-GYN-0803.
Study subjects or cohorts overlap
One study has been previously published on the same cohort:
Milon, A. et al Abbreviated breast MRI combining FAST protocol and high temporal resolution (HTR) dynamic contrast enhanced (DCE) sequence. European Journal of Radiology 117, 199–208 (2019).
Methodology
• retrospective
• diagnostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 1255 kb)
Rights and permissions
About this article
Cite this article
Perre, S.V., Duron, L., Milon, A. et al. Radiomic analysis of HTR-DCE MR sequences improves diagnostic performance compared to BI-RADS analysis of breast MR lesions. Eur Radiol 31, 4848–4859 (2021). https://doi.org/10.1007/s00330-020-07519-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07519-9