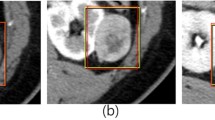
Abstract
Objectives
To develop a deep learning-based method for automated classification of renal cell carcinoma (RCC) from benign solid renal masses using contrast-enhanced computed tomography (CECT) images.
Methods
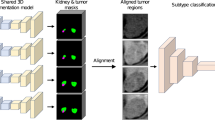
This institutional review board–approved retrospective study evaluated CECT in 315 patients with 77 benign (57 oncocytomas, and 20 fat-poor angiomyolipoma) and 238 malignant (RCC: 123 clear cell, 69 papillary, and 46 chromophobe subtypes) tumors identified consecutively between 2015 and 2017. We employed a decision fusion-based model to aggregate slice level predictions determined by convolutional neural network (CNN) via a majority voting system to evaluate renal masses on CECT. The CNN-based model was trained using 7023 slices with renal masses manually extracted from CECT images of 155 patients, cropped automatically around kidneys, and augmented artificially. We also examined the fully automated approach for renal mass evaluation on CECT. Moreover, a 3D CNN was trained and tested using the same datasets and the obtained results were compared with those acquired from slice-wise algorithms.
Results
For differentiation of RCC versus benign solid masses, the semi-automated majority voting-based CNN algorithm achieved accuracy, precision, and recall of 83.75%, 89.05%, and 91.73% using 160 test cases, respectively. Fully automated pipeline yielded accuracy, precision, and recall of 77.36%, 85.92%, and 87.22% on the same test cases, respectively. 3D CNN reported accuracy, precision, and recall of 79.24%, 90.32%, and 84.21% using 160 test cases, respectively.
Conclusions
A semi-automated majority voting CNN-based methodology enabled accurate classification of RCC from benign neoplasms among solid renal masses on CECT.
Key Points
• Our proposed semi-automated majority voting CNN-based algorithm achieved accuracy of 83.75% for the diagnosis of RCC from benign solid renal masses on CECT images.
• A fully automated CNN-based methodology classified solid renal masses with moderate accuracy of 77.36% using the same test images.
• Employing 3D CNN-based methodology yielded slightly lower accuracy for renal mass classification compared with the semi- automated 2D CNN-based algorithm (79.24%).





Similar content being viewed by others
Abbreviations
- AUC:
-
Area under curve
- CECT:
-
Contrast-enhanced computed tomography
- CNN:
-
Convolutional neural network
- DICOM:
-
Digital imaging and communication in medicine
- fpAML:
-
Fat-poor renal angiomyolipoma
- GPU:
-
Graphics processing unit
- LHIN:
-
Local health integration network
- PACS:
-
Picture archiving and communication system
- PPV:
-
Positive predictive value
- PR:
-
Precision-recall
- RCC:
-
Renal cell carcinoma
- ReLU:
-
Rectified linear unit
- ROI:
-
Region of interest
- ROC:
-
Receiver operating characteristic
- SD:
-
Standard deviation
References
Meyer HJ, Pfeil A, Schramm D, Bach AG, Surov A (2017) Renal incidental findings on computed tomography: frequency and distribution in a large non selected cohort. Medicine (Baltimore) 96:e7039–e7043
O’Connor SD, Pickhardt PJ, Kim DH, Oliva MR, Silverman SG (2011) Incidental finding of renal masses at unenhanced CT: prevalence and analysis of features for guiding management. AJR Am J Roentgenol 1:139–145
Schieda N, Lim RS, McInnes MDF et al (2018) Characterization of small (<4 cm) solid renal masses by computed tomography and magnetic resonance imaging: current evidence and further development. Diagn Interv Imaging 99:443–455
Lim CS, Schieda N, Silverman SG (2019) Update on indications for percutaneous renal mass biopsy in the era of advanced CT and MRI. AJR Am J Roentgenol 212:1187–1196
Udare A, Walker D, Krishna S et al (2019) Characterization of clear cell renal cell carcinoma and other renal tumors: evaluation of dual-energy CT using material-specific iodine and fat imaging. Eur Radiol. https://doi.org/10.1007/s00330-019-06590-1
Lee-Felker SA, Felker ER, Tan N et al (2014) Qualitative and quantitative MDCT features for differentiating clear cell renal cell carcinoma from other solid renal cortical masses. AJR Am J Roentgenol 203:W516–W524
Lim RS, Flood TA, McInnes MDF, Lavallee LT, Schieda N (2018) Renal angiomyolipoma without visible fat: can we make the diagnosis using CT and MRI? Eur Radiol 28:542–553
Kang SK, Huang WC, Pandharipande PV, Chandarana H (2014) Solid renal masses: what the numbers tell us. Am J Roentgenol 202:1196–1206
Schieda N, Al-Subhi M, Flood TA, El-Khodary M, McInnes MD (2014) Diagnostic accuracy of segmental enhancement inversion for the diagnosis of renal oncocytoma using biphasic computed tomography (CT) and multiphase contrast-enhanced magnetic resonance imaging (MRI). Eur Radiol 24:2787–2794
Schieda N, Al-Subhi M, Flood TA et al (2014) Diagnostic accuracy of segmental enhancement inversion for the diagnosis of renal oncocytoma using biphasic computed tomography (CT) and multiphase contrast-enhanced magnetic resonance imaging (MRI). Eur Radiol. https://doi.org/10.1007/s00330-014-3310-y
Hodgdon T, McInnes MDF, Schieda N, Flood TA, Lamb L, Thornhill RE (2015) Can quantitative CT texture analysis be used to differentiate fat-poor renal angiomyolipoma from renal cell carcinoma on unenhanced CT images? Radiology 276:787–796
Schieda N, Thornhill RE, Al-Subhi M et al (2015) Diagnosis of sarcomatoid renal cell carcinoma with CT: evaluation by qualitative imaging features and texture analysis. AJR Am J Roentgenol 204:1013–1023
Schieda N, Lim RS, Krishna S, McInnes MDF, Flood TA, Thornhill RE (2018) Diagnostic accuracy of unenhanced CT analysis to differentiate low-grade from high-grade chromophobe renal cell carcinoma. AJR Am J Roentgenol 210:1079–1087
Sasaguri K, Takahashi N, Gomez-Cardona D et al (2015) Small (< 4 cm) renal mass: differentiation of oncocytoma from renal cell carcinoma on biphasic contrast-enhanced CT. AJR Am J Roentgenol 205:999–1007
Raman SP, Chen Y, Schroeder JL, Huang P, Fishman EK (2014) CT texture analysis of renal masses: pilot study using random forest classification for prediction of pathology. Acad Radiol 21:1587–1596
Bektas CT, Kocak B, Yardimci AH et al (2019) Clear cell renal cell carcinoma: machine learning-based quantitative computed tomography texture analysis for prediction of Fuhrman nuclear grade. Eur Radiol 29:1153–1163
Kocak B, Yardimci AH, Bektas CT et al (2018) Textural differences between renal cell carcinoma subtypes: machine learning-based quantitative computed tomography texture analysis with independent external validation. Eur J Radiol 107:149–157
Feng Z, Rong P, Cao P et al (2018) Machine learning-based quantitative texture analysis of CT images of small renal masses: differentiation of angiomyolipoma without visible fat from renal cell carcinoma. Eur Radiol 28:1625–1633
Lee H, Hong H, Kim J, Jung DC (2018) Deep feature classification of angiomyolipoma without visible fat and renal cell carcinoma in abdominal contrast-enhanced CT images with texture image patches and hand-crafted feature concatenation. Med Phys 45:1550–1561
Lee HS, Hong H, Jung DC, Park S, Kim J (2017) Differentiation of fat-poor angiomyolipoma from clear cell renal cell carcinoma in contrast-enhanced MDCT images using quantitative feature classification. Med Phys 44:3604–3614
Coy H, Hsieh K, Wu W, Nagarajan MB, Young JR, Douek ML (2019) Deep learning and radiomics: the utility of Google TensorFlowTM inception in classifying clear cell renal cell carcinoma and oncocytoma on multiphasic CT. Abdom Radiol (NY) 44:2009–2020
Silverman SG, Pedrosa I, Ellis JH, Hindman NM, Schieda N, Smith AD (2017) Bosniak classification of cystic renal masses, version 2019: an update proposal and needs assessment. Radiology. 2019:292
Krishna S, Murray CA, McInnes MD, Chatelain R, Siddaiah M, Al-Dandan O, Narayanasamy S, Schieda N (2017) CT imaging of solid renal masses: pitfalls and solutions. Clin Radiol 72:708–721. https://doi.org/10.1016/j.crad.2017.05.003
Czarniecki M, Gautam R, Choyke PL, Turkbey B (2018) Imaging findings of hereditary renal tumors, a review of what the radiologist should know. Eur J Radiol 101:8–16
Moch H, Cubilla AL, Humphrey PA, Reuter VE, Ulbright TM (2016) The 2016 WHO classification of tumours of the urinary system and male genital organs—part a: renal, penile, and testicular tumours. Eur Urol 70:93–105
Zabihollahy F, Schieda N, Krishna S, Ukwatta E (2020) Ensemble U-Net-based method for fully automated detection and segmentation of renal masses on computed tomography images. J Med Phys. https://doi.org/10.1002/mp.14193
Saito T, Rehmsmeier M (2015) The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One. https://doi.org/10.1371/journal.pone.0118432
Hodgdon T, McInnes MDF, Schieda N et al (2015) Can quantitative CT texture analysis be used to differentiate fat-poor renal angiomyolipoma from renal cell carcinoma on unenhanced CT images? Radiology. https://doi.org/10.1148/radiol.2015142215
Zhou L, Zhang Z, Chen YC, Zhao ZY, Yin XD, Jiang HB (2019) A deep learning-based radiomics model for differentiating benign and malignant renal tumors. Transl Oncol 12:292–300
Acknowledgments
Fatemeh Zabihollahy acknowledges the Ontario Graduate Scholarship (OGS).
Funding
This study has received funding by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant (E. Ukwatta).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Dr. Nicola Schieda.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 218 kb)
Rights and permissions
About this article
Cite this article
Zabihollahy, F., Schieda, N., Krishna, S. et al. Automated classification of solid renal masses on contrast-enhanced computed tomography images using convolutional neural network with decision fusion. Eur Radiol 30, 5183–5190 (2020). https://doi.org/10.1007/s00330-020-06787-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-06787-9