Abstract
Key message
The overexpression of IbbZIP1 leads to a significant upregulation of abiotic-related genes, suggesting that IbbZIP1 gene confers salt and drought tolerance in transgenic Arabidopsis.
Abstract
Basic region/leucine zipper motif (bZIP) transcription factors regulate flower development, seed maturation, pathogen defense, and stress signaling in plants. Here, we cloned a novel bZIP transcription factor gene, named IbbZIP1, from sweetpotato [Ipomoea batatas (L.) Lam.] line HVB-3. The full length of IbbZIP1 exhibited transactivation activity in yeast. The expression of IbbZIP1 in sweetpotato was strongly induced by NaCl, PEG6000, and abscisic acid (ABA). Its overexpression in Arabidopsis significantly enhanced salt and drought tolerance. Under salt and drought stresses, the transgenic Arabidopsis plants showed significant upregulation of the genes involved in ABA and proline biosynthesis and reactive oxygen species scavenging system, significant increase of ABA and proline contents and superoxide dismutase activity and significant decrease of H2O2 content. These results demonstrate that the IbbZIP1 gene confers salt and drought tolerance in transgenic Arabidopsis. This study provides a novel bZIP gene for improving the tolerance of sweetpotato and other plants to abiotic stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salinity and drought seriously affect the productivity of agricultural crops in the world (Munns and Tester 2008; Yang et al. 2010; Zhao et al. 2013). Developing crops tolerant to salinity and drought is becoming important. Plants adapt to salinity and drought stresses by developing a variety of mechanisms, such as growth and development regulation, osmotic adjustment, detoxification, and ion homeostasis (Bohnert et al. 1995; Zhu 2001, 2002).
The basic region/leucine zipper motif (bZIP) proteins compose a large family of transcription factors in higher plants. The bZIP members have been reported to regulate flower development, seed maturation, pathogen defense and stress, light, hormone and sugar signaling pathways (Jakoby et al. 2002; Lindemose et al. 2013). Several bZIP genes have been found to confer the tolerance to abiotic stresses in some plant species. In Arabidopsis, AtABF3, AtbZIP24, and AtbZIP1 are positive regulators of plant tolerance to abiotic stresses (Kim et al. 2004; Yang et al. 2009; Sun et al. 2012). In rice, OsbZIP23, OsbZIP72, OsABF1/OsbZIP12, OsABF2/OsbZIP46, and OsbZIP71 increased the tolerance to abiotic stresses (Xiang et al. 2008; Lu et al. 2009; Hossain et al. 2010a, b; Liu et al. 2014a, b, c, d), but OsbZIP52 negatively regulates responses to cold and drought (Liu et al. 2012). GmbZIP44, GmbZIP62, and GmbZIP78 conferred salt and freezing tolerance in transgenic Arabidopsis (Liao et al. 2008). GmbZIP1 increased the tolerance to salinity, cold temperature and drought in transgenic Arabidopsis and improved drought tolerance in transgenic wheat (Gao et al. 2011a, b). ZmbZIP72 conferred salt and drought tolerance in transgenic Arabidopsis (Ying et al. 2012) and ZmABP9 enhanced salt and drought tolerance in transgenic cotton (Wang et al. 2017).
Sweetpotato is an important food crop worldwide (Zang et al. 2009). Its productivity is often limited by salinity and drought stresses (Liu et al. 2014a). Gene engineering is an alternative approach for improving abiotic stresses’ tolerance in sweetpotato (Kim et al. 2012, 2013, 2014; Liu et al. 2014a, b, c, 2015; Wang et al. 2016a, b, c; Zhai et al. 2015; Li et al. 2017). To date, the bZIP family has not been reported in sweetpotato. Here, a novel bZIP transcription factor gene, named IbbZIP1, was isolated from sweetpotato and found to be involved in salt and drought tolerance in transgenic Arabidopsis.
Materials and methods
Plant materials
Sweetpotato line HVB-3 was used to isolate the IbbZIP1 gene in this study. Its transcriptome sequencing was conducted by Li et al. (2017), from which one expressed sequence tag (EST) was obtained. Arabidopsis thaliana (Columbia-0, WT) was employed to analyze the function of IbbZIP1.
Cloning and analysis of IbbZIP1 and its promoter
Freshly-harvested storage roots of HVB-3 were used to extract total RNA. The first-strand cDNA was obtained using PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa, Beijing, China). A rapid amplification of cDNA ends (RACE) procedure was applied to amplify the full-length cDNA of IbbZIP1 with specific primers (Supplementary Table S1). Genomic DNA extracted from in vitro-grown plants was used to amplify the genomic sequence of IbbZIP1 (Wang et al. 2016a). Its promoter was cloned with Universal GenomeWalker 2.0 Kit (TaKaRa, Dalian, China). The specific primers listed in Supplementary Table S1 were employed.
The IbbZIP1 cDNA analysis was performed online (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) was used to predict its open-reading frame (ORF). The phylogenetic analysis was conducted with the DNAMAN software (Lynnon Biosoft, Quebec, Canada). Exon–intron structure was constructed using Splign tool (https://www.ncbi.nlm.nih.gov/sutils/splign/splign.cgi?textpage=online&level=form) and compared with the At1g58110 gene (http://www.arabidopsis.org/). The molecular weight and theoretical isoelectric point (pI) of the IbbZIP1 protein were analyzed online (http://web.expasy.org/compute_pi/). PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was employed to identify the cis-acting regulatory elements in the promoter region of IbbZIP1.
Expression analysis of IbbZIP1 in sweetpotato
Storage root, stem, and leaf tissues of HVB-3 grown for 100 days in a field were used to isolate total RNA. The expression of IbbZIP1 was analyzed by quantitative real-time PCR (qRT-PCR) with the specific primers of IbbZIP1 (Supplementary Table S1) and Ibactin (AY905538) as an internal control based on the method of Liu et al. (2014a). Comparative CT method was employed to quantify the gene expression (Schmittgen and Livak 2008). Furthermore, the HVB-3 plants cultured for 4 weeks on the basal Murashige and Skoog (MS) medium were transferred to liquid MS medium with H2O (control), 200 mM NaCl, 20% PEG6000 and 100 μM ABA, respectively, and sampled at 0, 2, 4, 6, 12, 24, and 48 h after treatment for analyzing the expression of IbbZIP1.
Transactivation assay of IbbZIP1 in yeast
Transactivation assay of the IbbZIP1 protein in yeast (Saccharomyces cerevisiae) was done as described by Jiang et al. (2014). Its encoding region obtained with specific primers (Supplementary Table S1) was integrated into the vector pGBKT7 (pBD). Expression vector pBD-IbbZIP1, pGAL4 (as positive vector) and pBD (as negative vector) were transferred into the yeast strain AH109, respectively. The transactivation activity was determined according to the method of Wang et al. (2016a).
Production of the transgenic Arabidopsis plants
The overexpression vector pC3301-121-IbbZIP1 was constructed by inserting 35S-IbbZIP1-NOS into pCAMBIA3301 at HindIII and EcoRI and then transferred into the Agrobacterium tumefaciens strain GV3101. The dipping flower method was applied to transform Arabidopsis (Clough and Bent 1998). The transgenic Arabidopsis plants were determined by histochemical GUS assay (Jefferson et al. 1987) and PCR analysis with 35S forward and IbbZIP1-specific reverse primers (Supplementary Table S1).
Assay for salt and drought tolerance
Transgenic Arabidopsis T3 and WT seedlings were treated on MS medium containing 200 mM NaCl or 300 mM mannitol under 13 h day-light at 54 μM/m2/s and 22 °C. After 2 weeks, their root length and fresh weight were investigated. Furthermore, the T3 and WT seedlings were planted for 2 weeks in pots with a soil, vermiculite, and humus mixture (1:1:1, v/v/v) and each pot was irrigated with a 33 mL of 300 mM NaCl solution once every 2 days for 2 weeks, or stressed by drought for 4 weeks and re-watered for 2 days. The contents of abscisic acid (ABA) and proline and the activity of superoxide dismutase (SOD) in the T3 and WT plants treated in pots for 4 weeks under no stresses, 1 week under 300 mM NaCl stress or 2 weeks under drought stress were determined as described by Gao et al. (2011a). The H2O2 content was measured with H2O2 Assay Kit (Comin Biotechnology Co., Ltd. Suzhou, China). Twenty-seven plants in three pots with nine plants per pot were treated for each line.
Assay for ABA sensitivity
For ABA sensitivity assay, the transgenic Arabidopsis and WT seeds (50 seeds for each line) were sown on MS medium with 0, 0.5, and 1 μM ABA, respectively, under 13 h day-light at 54 μM/m2/s and 22 °C. After 1 week, their germination rate and cotyledon opening and greening rate were investigated.
Expression analysis of the related genes
The T3 and WT plants potted for 4 weeks without stress, 1 week stressed with 300 mM NaCl or 2 weeks stressed by drought were employed to analyze the expression of the genes involved in ABA and proline biosynthesis and reactive oxygen species (ROS) scavenging system using qRT-PCR protocols of Liu et al. (2014a). The specific primers of Atactin (internal control, NM112764) and the related genes were listed in Supplementary Table S1.
Statistical analysis
All experiments were performed with three biological replicates. Data presented as the mean ± SE were analyzed by Student’s t test (two-tailed analysis) at P < 0.05 (*) and P < 0.01 (**).
Results
Cloning and sequence analysis of IbbZIP1 and its promoter
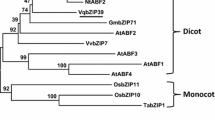
The IbbZIP1 gene was cloned from sweetpotato line HVB-3. The cDNA sequence was 1757 bp in length and contained an 1080 bp ORF encoding a 359-aa polypeptide with a molecular weight of 41.26 kDa and a predicted pI of 7.89. The IbbZIP1 protein was subjected to phylogenetic analysis together with 44 Arabidopsis bZIP proteins belonging to 10 groups (Jakoby et al. 2002). The results revealed that IbbZIP1 belonged to group E of the bZIP family and had a close relationship with At1g58110 (Fig. 1). IbbZIP1 contained one bZIP domain (Fig. 2a). The 1946 bp genomic DNA of IbbZIP1 contained 4 exons and 3 introns (Fig. 2b). Its promoter region (~ 2056 bp) had the stress-responsive cis-acting regulatory elements, such as HSE, MBS, TCA, GARE, TGA, ERE, and ABRE (Supplementary Fig. S1; Supplementary Table S2).
Phylogenetic analysis of IbbZIP1 with 44 Arabidopsis bZIP proteins. Ten groups of bZIP proteins were defined based on the classification of Arabidopsis bZIP proteins (Jakoby et al. 2002)
Comparison between IbbZIP1 and the Arabidopsis E group members. a Sequence alignment of IbbZIP1 with Arabidopsis proteins belonging to the E group. Black line shows the basic leucine zipper (bZIP) domain. b Comparison of exon and intron constituents between IbbZIP1 and At1g58110. Exons are represented by colorful boxes and introns by black lines, with length (bp) displayed above exons and below introns
Expression of IbbZIP1 in sweetpotato
The IbbZIP1 gene exhibited significantly higher expression level in the leaves of HVB-3 than in the stems and storage roots (Fig. 3a). Its expression in the in vitro-grown plants of HVB-3 was strongly induced by NaCl, PEG6000 and ABA, and peaked (6.79-fold) at 12 h of 200 mM NaCl treatment, (6.58-fold) at 24 h of 20% PEG6000 treatment and (7.51-fold) at 24 h of 100 μM ABA treatment (Fig. 3b). These results showed that IbbZIP1 might be involved in the tolerance to salt and drought in sweetpotato.
a Expression analysis of IbbZIP1 in storage root, stem and leaf tissues of HVB-3. Different capital letters indicate a significant difference at P < 0.01 by Student’s t test. b Expression analysis of the IbbZIP1 gene in the in vitro-grown plants of HVB-3 after different times (h) in response to H2O, 200 mM NaCl, 20% PEG6000 and 100 μM ABA, respectively. Data are presented as mean values ± SE (n = 3)
Transactivation activity of IbbZIP1 in yeast
The yeast one-hybrid system was applied to identify a possible transactivation activity of the IbbZIP1 protein. The yeast cells harboring pGAL4 and pGBKT7-IbbZIP1 grew well on SD plate without tryptophan and histidine and exhibited β-galactosidase activity, but the cells bearing pBD failed to grow (Fig. 4). These results demonstrated that IbbZIP1 had transactivation activity in yeast and has potential ability to activate the transcription of related genes by directly binding to cis-acting elements in their promoter region or interacting with other proteins.
Production of the transgenic Arabidopsis plants
Lots of putatively transgenic Arabidopsis seeds were obtained by the dipping flower method. The randomly sampled seeds were sown on MS medium with 12.5 mg/L phosphinothricin (PPT) and produced plants. GUS assay and PCR analysis confirmed that 8 of the randomly sampled 60 plants were transgenic plants, named L1, L2, …, L8, respectively (Supplementary Fig. S2), from which T3 were generated. The expression of IbbZIP1 in the leaves of the T3 and WT plants was analyzed by qRT-PCR and 4 of them (L1, L3, L6 and L8) exhibited significantly higher expression level compared with other plants (Supplementary Fig. S2).
Enhanced salt and drought tolerance of transgenic Arabidopsis
Four transgenic lines (L1, L3, L6, and L8) and WT seedlings were cultured on MS medium with 200 mM NaCl, 300 mM mannitol or without stresses for 2 weeks, respectively. The transgenic and WT plants exhibited no differences in growth without stresses, but the transgenic plants showed significantly better growth and increased physical size than WT under NaCl and mannitol stresses (Fig. 5).
Responses of the transgenic Arabidopsis seedlings and WT cultured for 2 weeks on MS medium with 200 mM NaCl and 300 mM mannitol, respectively. a Growth and rooting of the transgenic Arabidopsis seedlings and WT. b Root length and plant fresh weight of the transgenic Arabidopsis seedlings and WT. Data are presented as mean ± SE (n = 3)
Two-week-old transgenic and WT plants grown in pots were stressed by 300 mM NaCl and drought, respectively. The transgenic plants and WT showed no differences in growth without stresses (Fig. 6a). Under salt and drought stresses, the transgenic plants exhibited good growth, increased ABA and proline contents, enhanced SOD activity and decreased H2O2 content, while WT almost died (Fig. 6a, b). These results indicated that L1, L3, L6, and L8 had significantly enhanced salt and drought tolerance compared with WT.
Responses of the transgenic Arabidopsis plants and WT grown in pots under salt and drought stresses. a Phenotypes of transgenic vs. WT plants grown for 6 weeks under normal condition, 2 weeks under 300 mM NaCl stress and 4 weeks under drought stress and then 2 days after re-watering, respectively. b ABA, proline, and H2O2 contents and SOD activity in the transgenic plants and WT grown for 4 weeks under normal condition, 1 weeks under 300 mM NaCl and 2 weeks under drought stress. Data are presented as mean ± SE (n = 3)
Increased ABA sensitivity of transgenic Arabidopsis
Seed germination in response to ABA was tested to determine whether IbbZIP1 is involved in the ABA signaling pathway. No obvious differences in germination rate and cotyledon opening and greening rate were observed between the transgenic lines (L1, L3, L6, and L8) and WT under normal condition (Fig. 7). With exposure to different concentrations of ABA, both germination rate and cotyledon opening and greening rate of the transgenic and WT seeds declined, but the germination of L1, L3, L6, and L8 seeds were more sensitive to ABA-elicited inhibition, indicating that this gene might be involved in the ABA signaling pathway (Fig. 7).
Responses of the transgenic Arabidopsis seeds and WT sown on MS medium with 0, 0.5, and 1 μM ABA for 1 week. a Growth vigor of the transgenic and WT seedlings. b Germination rates of the transgenic Arabidopsis seeds and WT. c Cotyledon opening and greening rates of the transgenic Arabidopsis seeds and WT
Expression of the stress-responsive genes in the transgenic Arabidopsis plants
The genes encoding ABA biosynthetic 9-cis-epoxycarotenoid dioxygenase (NCED) and xanthoxin dehydrogenase (ABA2) and proline biosynthetic pyrroline-5-carboxylate synthase (P5CS) showed significantly higher expression levels in the transgenic plants in comparison with WT under salt and drought stresses (Fig. 8). The ROS scavenging genes encoding SOD, glutathione peroxidase (GPX), catalase (CAT), ascorbate peroxidase (APX) and dehydroascorbate reductase (DHAR) were found to show a systematic upregulation in the transgenic plants under salt and drought stresses (Fig. 8).
Transcript levels of salt- and drought-responsive genes in the transgenic Arabidopsis plants and WT under salt and drought stresses. Leaves of transgenic and WT pot-grown plants incubated for 4 weeks under normal condition, 2 weeks under 300 mM NaCl and 4 weeks under drought stress were used for expression analysis of the genes. Data are presented as mean ± SE (n = 3)
Discussion
Several bZIP transcription factor genes have been cloned from Arabidopsis, rice, soybean, maize etc., and found to play roles in plant stress-responsive and hormone signal transduction (Sun et al. 2012; Liu et al. 2014a; Gao et al. 2011a; Wang et al. 2017). However, there is no report on the bZIP transcription factors in sweetpotato. This study reported, for the first time, the cloning and characterization of a novel transcription factor gene, IbbZIP1, from sweetpotato. The IbbZIP1 protein contains a typical bZIP domain which is essential for bZIP transcription factor, belongs to E group of bZIP family and has a close relationship with At1g58110 (Figs. 1, 2). To date, the function of the At1g58110 gene is still unknown. We found that the promoter region of IbbZIP1 had the stress-responsive cis-acting regulatory elements (Yamaguchi-Shinozaki et al. 2005, 2006; Xiang et al. 2008). The IbbZIP1 expression was induced by NaCl, PEG, and ABA (Figs. S1, 4; Supplementary Table S2) and its overexpression increased salt and drought tolerance in transgenic Arabidopsis (Figs. 6, 7).
The bZIP transcription factors regulate the ABA-mediated abiotic stresses signaling pathways in plants (Mehrotra et al. 2014). In rice, overexpression of OsABI5 and OsbZIP23 upregulated the ABA biosynthetic gene ABA2 (Zou et al. 2008; Xiang et al. 2008). The ABA biosynthetic gene NCED2 was upregulated in the ZmABP9-overexpressing cotton plants (Wang et al. 2017). ABA regulates the expression of ABA dependent stress-responsive genes and higher levels of ABA can enhance salt and drought tolerance in Arabidopsis (Tuteja 2007; Liu et al. 2013; Wang et al. 2016a). In the present study, increased ABA sensitivity of transgenic Arabidopsis to exogenous ABA showed that IbbZIP1 might be involved in the ABA signaling pathway (Fig. 7). The IbbZIP1-overexpressing Arabidopsis plants exhibited significant upregulation of NCED and ABA2 and significant increase of ABA content under salt and drought stresses (Figs. 7, 8). It is suggested that overexpression of IbbZIP1 confers tolerance to salt and drought due to the increased expression level of the ABA biosynthetic genes, which increases the production of ABA as a signaling molecule and further the expression of stress-responsive genes.
It has been reported that the high level of ABA in rice increases the transcript level of OsP5CS1, which lead to accumulation of proline under abiotic stresses (Sripinyowanich et al. 2013). It has been shown that proline accumulation resulted in the enhanced salt and drought tolerance in several plant species (Szabados and Savouré 2010; Krasensky and Jonak 2012; Zhang et al. 2012; Liu et al. 2015). Overexpression of GmbZIP62 and GmbZIP78 upregulated the genes involved in ABA signaling pathways and GmP5CS1, resulting the increased proline content and the enhanced salt and freezing tolerance in transgenic Arabidopsis (Liao et al. 2008). In this study, the IbbZIP1-overexpressing Arabidopsis plants exhibited the increased P5CS expression level and proline content under salt and drought stresses (Figs. 7, 8). These results suggested that more proline might be accumulated because of the increased ABA level, which leads to the enhanced salt and drought tolerance.
In plants, overproduction of ROS often occurs under salinity and drought stresses, which leads to oxidative damage. ROS can be detoxified by activating ROS scavenging enzymes (Gill and Tuteja 2010). The increased proline level led to upregulation of the ROS scavenging genes, which resulted in the enhanced tolerance to salt and drought in transgenic sweetpotato (Liu et al. 2014b, Liu et al. 2015; Wang et al. 2016b; Zhai et al. 2015). We found that under salt and drought stresses, the ROS scavenging genes (SOD, GPX, CAT, APX, and DHAR) were systematically upregulated in the IbbZIP1-overexpressing Arabidopsis plants (Fig. 8). Thus, more accumulation of proline in the IbbZIP1-overexpressing Arabidopsis plants might upregulate the ROS scavenging genes and further stimulate the ROS scavenging system, which results in the enhanced salt and drought tolerance.
In conclusion, a novel sweetpotato bZIP transcription factor gene, IbbZIP1, has been successfully isolated. The IbbZIP1 gene is involved in salt and drought tolerance in transgenic Arabidopsis. Its overexpression might upregulate the genes involved in ABA and proline biosynthesis and ROS scavenging system and increase the ABA and proline contents, which leads to enhanced salt and drought tolerance. This study provides a novel bZIP gene for improving the tolerance of sweetpotato and other plants to abiotic stresses.
Author contribution statement
Conceived and designed the experiments: QCL CK. Performed the experiments: CK. Analyzed the data: CK. Contributed reagents/materials/analysis tools: QCL HZ SZH NZ. Wrote the paper: QCL CK.
Abbreviations
- bZIP:
-
Basic region/leucine zipper motif
- EST:
-
Expressed sequence tag
- RACE:
-
Rapid amplification of cDNA ends
- pI:
-
Isoelectric point
- ABA:
-
Abscisic acid
- SOD:
-
Superoxide dismutase
- ROS:
-
Reactive oxygen species
- PPT:
-
Phosphinothricin
References
Bohnert HJ, Nelson DE, Jensen RG (1995) Adaptations to environmental stresses. Plant Cell 7:1099–1111
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Gao S, Yuan L, Zhai H, Liu CL, He SZ, Liu QC (2011a) Transgenic sweetpotato plants expressing an LOS5 gene are tolerant to salt stress. Plant Cell, Tissue Organ Cult 107:205–213
Gao SQ, Chen M, Xu ZS, Zhao CP, Li LC, Xu HJ, Tang YM, Zhao X, Ma YZ (2011b) The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol Biol 75:537–553
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930
Hossain MA, Cho JI, Han M, Ahn CH, Jeon JS, An G, Park PB (2010a) The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J Plant Physiol 167:1512–1520
Hossain MA, Lee Y, Cho JI, Ahn CH, Lee SK, Jeon JS, Kang H, Lee CH, An G, Park PB (2010b) The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol 72:557–566
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111
Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Jiang XQ, Zhang CQ, Lü PT, Jiang GM, Liu XW, Dai FW, Gao JP (2014) RhNAC3, a stress-associated NAC transcription factor, has a role in dehydration tolerance through regulating osmotic stress-related genes in rose petals. Plant Biotechnol J 12:38–48
Kim JB, Kang JY, Kim SY (2004) Over-expression of a transcription factor regulating ABA-responsive gene expression confers multiple stress tolerance. Plant Biotechnol J 2:459–466
Kim SH, Ahn YO, Ahn MJ, Lee HS, Kwak SS (2012) Down-regulation of β-carotene hydroxylase increases β-carotene and total carotenoids enhancing salt stress tolerance in transgenic cultured cells of sweetpotato. Phytochemistry 74:69–78
Kim SH, Kim YH, Ahn YO, Ahn MJ, Jeong JC, Lee HS, Kwak SS (2013) Downregulation of the lycopene ε-cyclase gene increases carotenoid synthesis via the β-branch-specific pathway and enhances salt-stress tolerance in sweetpotato transgenic calli. Physiol Plant 147:432–442
Kim SH, Jeong JC, Park S, Bae JY, Ahn MJ, Lee HS, Kwak SS (2014) Down-regulation of sweetpotato lycopene β-cyclase gene enhances tolerance to abiotic stress in transgenic calli. Mol Biol Rep 41:8137–8148
Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63:1593–1608
Li RJ, Kang C, Song XJ, Yu L, Liu DG, He SZ, Zhai H, Liu QC (2017) A ζ-carotene desaturase gene, IbZDS, increases β-carotene and lutein contents and enhances salt tolerance in transgenic sweetpotato. Plant Sci 262:39–51
Liao Y, Zou H, Wei W, Hao YJ, Tian AG, Huang J, Liu YF, Zhang JS, Chen SY (2008) Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228:225–240
Lindemose S, O’Shea C, Jensen MK, Skriver K (2013) Structure, function and networks of transcription factors involved in abiotic stress responses. Int J Mol Sci 14:5842–5878
Liu CT, Wu YB, Wang XP (2012) bZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice. Planta 235:1157–1169
Liu XX, Zhai SM, Zhao YJ, Sun BC, Liu C, Yang AF, Zhang JR (2013) Overexpression of the phosphatidylinositol synthase gene (ZmPIS) conferring drought stress tolerance by altering membrane lipid composition and increasing ABA synthesis in maize. Plant, Cell Environ 36:1037–1055
Liu CT, Mao BG, Ou SJ, Wang W, Liu LC, Wu YB, Chu CC, Wang XP (2014a) OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol 84:19–36
Liu DG, He SZ, Zhai H, Wang LJ, Zhao Y, Wang B, Li RJ, Liu QC (2014b) Overexpression of IbP5CR enhances salt tolerance in transgenic sweetpotato. Plant Cell, Tissue Organ Cult 117:1–16
Liu DG, Wang LJ, Liu CL, Song XJ, He SZ, Zhai H, Liu QC (2014c) An Ipomoea batatas iron-sulfur cluster scaffold protein gene, IbNFU1, is involved in salt tolerance. PLoS ONE 9:e93935
Liu DG, Wang LJ, Zhai H, Song XJ, He SZ, Liu QC (2014d) A novel a/b-hydrolase gene IbMas enhances salt tolerance in transgenic sweetpotato. PLoS ONE 9:e115128
Liu DG, He SZ, Song XJ, Zhai H, Liu N, Zhang DD, Ren ZT, Liu QC (2015) IbSIMT1, a novel salt-induced methyltransferase gene from Ipomoea batatas, is involved in salt tolerance. Plant Cell, Tissue Organ Cult 120:701–715
Lu GJ, Gao CX, Zheng XN, Han B (2009) Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229:605–615
Mehrotra R, Bhalothia P, Bansal P, Basantani MK, Bharti V, Mehrotra S (2014) Abscisic acid and abiotic stress tolerance—different tiers of regulation. J Plant Physiol 171:486–496
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C T method. Nat Protoc 3:1101–1108
Sripinyowanich S, Klomsakul P, Boonburapong B, Bangyeekhun T, Asami T, Gu HY, Buaboocha T, Chadchawan S (2013) Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): the role of OsP5CS1 and OsP5CR gene expression during salt stress. Environ Exp Bot 86:94–105
Sun XL, Li Y, Cai H, Bai X, Ji W, Ding XD, Zhu YM (2012) The Arabidopsis AtbZIP1 transcription factor is a positive regulator of plant tolerance to salt, osmotic and drought stresses. J Plant Res 125:429–438
Szabados L, Savouré A (2010) Proline: a multifunctional amino acid. Trends Plant Sci 15:89–97
Tuteja N (2007) Abscisic acid and abiotic stress signaling. Plant Signal Behav 2:135–138
Wang B, Zhai H, He SZ, Zhang H, Ren ZT, Zhang DD, Liu QC (2016a) A vacuolar Na +/H + antiporter gene, IbNHX2, enhances salt and drought tolerance in transgenic sweetpotato. Sci Hortic 201:153–166
Wang FB, Tong WJ, Zhu H, Kong WL, Peng RH, Liu QC, Yao QH (2016b) A novel Cys2/His2 zinc finger protein gene from sweetpotato, IbZFP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Planta 243:783–797
Wang FB, Zhai H, An YY, Si ZZ, He SZ, Liu QC (2016c) Overexpression of IbMIPS1 gene enhances salt tolerance in transgenic sweetpotato. J Integr Agric 15:271–281
Wang CL, Lu GQ, Hao YQ, Guo HM, Guo Y, Zhao J, Cheng HM (2017) ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 246:453–469
Xiang Y, Tang N, Du H, Ye H, Xiong LZ (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148:1938–1952
Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10:88–94
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Ann Rev Plant Biol 57:781–803
Yang O, Popova OV, Süthoff U, Lüking I, Dietz K, Golldack D (2009) The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 436:45–55
Yang SJ, Vanderbeld B, Wan JX, Huang YF (2010) Narrowing down the targets: towards successful genetic engineering of drought-tolerant crops. Mol Plant 3:469–490
Ying S, Zhang DF, Fu J, Shi YS, Song YC, Wang TY, Li Y (2012) Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 235:453–469
Zang N, Zhai H, Gao S, Chen W, He SZ, Liu QC (2009) Efficient production of transgenic plants using the bar gene for herbicide resistance in sweetpotato. Sci Hortic 122:649–653
Zhai H, Wang FB, Si ZZ, Huo JX, Xing L, An YY, He SZ, Liu QC (2015) A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol J 14:592–602
Zhang H, Han B, Wang T, Chen SX, Li HY, Zhang YH, Dai SJ (2012) Mechanisms of plant salt response: insights from proteomics. J Proteome Res 11:49–67
Zhao Q, Zhang H, Wang T, Chen S, Dai SJ (2013) Proteomics-based investigation of salt-responsive mechanisms in plant roots. J Proteoeme 82:230–253
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zou MJ, Guan YC, Ren HB, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66:675–683
Acknowledgements
This work was supported by National Natural Science Foundation of China (31461143017) and China Agriculture Research System (CARS-10, Sweetpotato).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Sang-Soo Kwak.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kang, C., Zhai, H., He, S. et al. A novel sweetpotato bZIP transcription factor gene, IbbZIP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Plant Cell Rep 38, 1373–1382 (2019). https://doi.org/10.1007/s00299-019-02441-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02441-x