Abstract
Key message
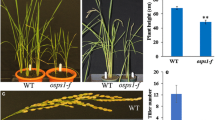
Targeted mutations in five carotenoid catabolism genes failed to boost carotenoid accumulation in rice seeds, but produced dwarf and high tillering mutants when OsCCD7 gene was knocked out.
Abstract
Carotenoids play an important role in human diet as a source of vitamin A. Rice is a major staple food in Asia, but does not accumulate carotenoids in the endosperm because of the low carotenoid biosynthesis or the degradation in metabolism. In this study, the CRISPR/Cas9 system was investigated in the targeted knockout of five rice carotenoid catabolic genes (OsCYP97A4, OsDSM2, OsCCD4a, OsCCD4b and OsCCD7) and in an effort to increase β-carotene accumulation in rice endosperm. Transgenic plants that expressed OsNLSCas9 and sgRNAs were generated by Agrobacterium-mediated transformation. Various knockout mutations were identified at the T0 generation of the transgenic rice by TILLING and direct sequencing of the PCR products amplified from the target sites. Carotenoids were not accumulated in both mono-allelic and bi-allelic knockout mutations of the five genes. However, transgenic plants with homozygous or bi-allelic mutations to the OsCCD7 gene were extremely dwarfish with more tillers and lower seed setting than other transgenic or nontransgenic plants. This phenotype was similar to the previously reported ccd7 mutants, which are defective in the biosynthesis of strigolactone, a plant hormone that regulates branching in plants and tiller formation in rice.








Similar content being viewed by others
Abbreviations
- 2,4-D:
-
2,4-Dichlorophenoxyacetic acid
- 6-BA:
-
6-Benzylaminopurine
- ABA:
-
Abscisic acid
- CCD:
-
Carotenoid cleavage dioxygenase
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- Cas9:
-
CRISPR-associated protein 9
- crRNA:
-
CRISPR RNA
- DNA:
-
Deoxyribonucleic acid
- EB:
-
Ethidium bromide
- EDTA:
-
Ethylene diamine tetraacetic acid
- HPTII:
-
Hygromycin phosphotransferase II gene
- NAA:
-
1-Naphthaleneacetic acid
- NCED:
-
Nine-cis-epoxy carotenoid dioxygenase
- NLS:
-
Nulcear localization signal
- PAM:
-
Protospacer-adjacent motif
- PCR:
-
Polymerase chain reaction
- PSY:
-
Phytoene synthase
- RNA:
-
Ribonucleic acid
- RT-PCR:
-
Reverse transcription PCR
- SDS:
-
Sodium dodecyl sulfate
- sgRNA:
-
Single guide RNA
- SSR:
-
Site-specific recombination
- TALEN:
-
Transcription activator-like effector nuclease
- tracrRNA:
-
Trans-activating crRNA
- WT:
-
Wild type
- ZFN:
-
Zinc finger nuclease
References
Burkhardt PK, Beyer P, Wünn J, Klöti A, Armstrong GA, Schledz M, von Lintig J, Potrykus I (1997) Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin A biosynthesis. Plant J 11:1071–1078
Cao Y, Wang J, Guo L, Xiao K (2008) Identification, characterization and expression analysis of transcription factor (CBF) genes in rice (Oryza sativa L.). Front Agric China 2:253–261
Cardoso C, Zhang Y, Jamil M, Hepworth J, Charnikhova T, Dimkpa SON, Meharg C, Wright MH, Liu J, Meng X, Wang Y, Li J, McCouch SR, Leyser O, Price AH, Bouwmeester HJ, Ruyter-Spira C (2014) Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1 orthologs. PNAS 111:2379–2384
Du H, Wang NL, Cui F, Li XH, Xiao JH, Xiong LZ (2010) Characterization of the β-carotene hydroxylase gene dsm2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant Physiol 154:1304–1318
Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19:1349
Fischer RA, Kohn GD (1966) The relationship of grain yield to vegetative growth and post-flowering leaf area in the wheat crop under conditions of limited soil moisture. Aust J Agric Res 17:281–295
Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826
Gautier H, Guichard S, Tchamitchian M (2001) Modulation of competition between fruits and leaves by flower pruning and water fogging, and consequences on tomato leaf and fruit growth. Ann Bot 88:645–652
Gonzalez-Jorge S, Ha SH, Magallanes-Lundback M, Gilliland LU, Zhou A, Lipka AE, Nguyen YN, Angelovici R, Lin H, Cepela J, Little H, Buell CR, Gore MA, Dellapenna D (2013) Carotenoid cleavage dioxygenase4 is a negative regulator of β-carotene content in Arabidopsis seeds. Plant Cell 25:4812–4826
Hikosaka S, Sugiyama N (2003) Fruit growth patterns and abortion in multi-pistillate type cucumbers. J Hortic Sci Biotechnol 78:775–779
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31:827–832
Ilg A, Yu QJ, Schaub P, Beyer P, Al-Babili S (2010) Overexpression of the rice carotenoid cleavage dioxygenase 1 gene in Golden Rice endosperm suggests apocarotenoids as substrates in planta. Planta 232:691–699
Jacobs TB, LaFayette PR, Schmitz JR, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 15:16
Kulkarni KP, Vishwakarma C, Sahoo SP, Lima JM, Nath M, Dokku P, Gacche RN, Mohapatra T, Robin S, Sarla N, Seshashayee M, Singh AK, Singh K, Singh NK, Sharma RP (2014) A substitution mutation in OsCCD7 cosegregates with dwarf and increased tillering phenotype in rice. J Genet 93:389–401
Lv MZ, Chao DY, Shan JX, Zhu MZ, Shi M, Gao JP, Lin HX (2012) Rice carotenoid b-ring hydroxylase CYP97A4 is involved in lutein biosynthesis. Plant Cell Physiol 53:987–1002
Marcelis LFM, Heuvelink E, Baan Hofman-Eijer LR, Den Bakker J, Xue LB (2004) Flower and fruit abortion in sweet pepper in relation to source and sink strength. J Exp Bot 55:2261–2268
Messing SA, Gabelli SB, Echeverria I, Vogel JT, Guan JC, Tan BC, Klee HJ, McCarty DR, Amzel LM (2010) Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 22:2970–2980
Nisar N, Li L, Lu S, Khin NC, Pogson BJ (2015) Carotenoid metabolism in plants. Mol Plant 8:68–82
Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR (2013) High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31:839–843
Schaub P, Al-Babili S, Drake R, Beyer P (2005) Why is golden rice golden (yellow) instead of red? Plant Physiol 138:441–450
Till B, Zerr T, Comai L, Henikoff S (2006) A protocol for TILLING and Ecotilling in plants and animals. Nat Protoc 1:2465–2477
Umehara M, Hanada A, Magome H, Takeda-Kamiya N, Yamaguchi S (2010) Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol 51:1118–1126
Vallabhaneni R, Bradbury LMT, Wurtzel ET (2010) The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch Biochem Biophys 504:104–111
Vogel JT, Tan BC, McCarty DR, Klee HJ (2008) The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J Biol Chem 283:11364–11373
Wang ZP, Xing HL, Dong L, Zhang HY, Han CY, Wang XC, Chen QJ (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol 16:144
Welsch R, Wust F, Bar C, Al-Babili S, Beyer P (2008) A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol 147:367–380
Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287:303–305
Yeum KJ, Russell RM (2002) Carotenoid bioavailability and bioconversion. Annu Rev Nutr 22:483–504
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu JK (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42:10903–10914
Zou J, Chen Z, Zhang S, Zhang W, Jiang G, Zhao X, Zhai W, Pan X, Zhu L (2005) Characterizations and fine mapping of a mutant gene for high tillering and dwarf in rice (Oryza sativa L.). Planta 222:604–612
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (Project No. 31271422) to WY, Shenzhen Peacock Innovation Team Project (No. KQTD201101) to WY and JH, Guangdong Innovation Research Team Fund (2014ZT05S078) to WY, Natural Science Foundation of Guangdong Province (Project No. 2015A030310421) to LC, and Hong Kong RGC AoE Project AoE/M-05/12 to JH. We thank Chao Sun, Meiting Wu, Linlin Luo and Ye Jin for assistance in analyzing the off-target cleavage activity.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Kan Wang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, X., Chen, L., He, J. et al. Knocking out of carotenoid catabolic genes in rice fails to boost carotenoid accumulation, but reveals a mutation in strigolactone biosynthesis. Plant Cell Rep 36, 1533–1545 (2017). https://doi.org/10.1007/s00299-017-2172-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-017-2172-6