Abstract
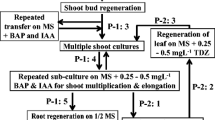
DNA methylation can control gene expression and may also play a role in plant development. Methylation of cytosine residues in DNA is enzymatically catalyzed by DNA methyltransferases. In this study, full-length genomic genes and cDNAs of methyltransferase (MET1) and domain-rearranged methyltransferase (DRM) were isolated from strawberry (Fragaria × ananassa Duch.). Two genomic clones (FaMET1a and FaMET1b) encoding MET1 had open-reading frame of 4,695 and 4,671 nucleotides with two introns, respectively. Amino acid sequence comparison indicated high similarity (98.72% identity) of strawberry MET1 protein to other plant MET1 sequences. The full-length cDNA of strawberry DRM genes (FaDRMa, FaDRMb and FaDRMc) were 2,273, 2,282 and 2,288 bp, respectively. Ten introns with different sizes were dispersed in FaDRM genes. Similarly, FaDRMa, FaDRMb and FaDRMc had high-sequence similarity overall. Expressions of strawberry MET1 and DRM genes were compared among in vitro-micropropagated plants, generations of micropropagated plants and conventionally propagated plants. The transcriptional expressions of both FaMET1 and FaDRM genes were downregulated in micropropagated plants, and they were recovered in the first and second runner generations of micropropagated plants. However, there was a slighter difference in global DNA methylation rates between micropropagated plants and conventionally propagated plants. Therefore, there was no positive relation between global DNA methylation rates and the expression levels of MET1 and DRM genes.









Similar content being viewed by others
Abbreviations
- IBA:
-
Indole-3-butyric acid
- MET:
-
Methyltransferase
- DRM:
-
Domain-rearranged methyltransferase
- DMT:
-
DNA-(cytosine-5) methyltransferase
- HPLC:
-
High-performance liquid chromatography
References
Ashapkin VV, Kutueva LI, Vanyushin BF (2002) The gene for domains rearranged methyltransferase (DRM2) in Arabidopsis thaliana plants is methylated at both cytosine and adenine residues. FEBS Lett 532(3):367–372
Ashley MV, Wilk JA, Styan SM, Craft KJ, Jones KL, Feldheim KA, Lewers KS, Ashman TL (2003) High variability and disomic segregation of microsatellites in the octoploid Fragaria virginiana Mill. (Rosaceae). Theor Appl Genet 107(7):1201–1207
Aufsatz W, Mette MF, Matzke AJ, Matzke M (2004) The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Mol Biol 54(6):793–804
Bartee L, Malagnac F, Bender J (2001) Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev 15(14):1753–1758
Bender J (2004) DNA methylation and epigenetics. Annu Rev Plant Biol 55:41–68
Bernacchia G, Para A, Pedrali-Noy G, Cella R (1998a) Isolation of a cDNA coding for DNA (cytosine-5)-methyltransferase (accession no. AJ002140) from Lycopersicon esculentum. Plant Physiol 116:446–446
Bernacchia G, Primo A, Giorgetti L, Pitto L, Cella R (1998b) Carrot DNA-methyltransferase is encoded by two classes of genes with differing patterns of expression. Plant J 13(3):317–329
Bitonti MB, Cozza R, Chiappetta A, Giannino D, Ruffini Castiglione M, Dewitte W, Mariotti D, Van Onckelen H, Innocenti AM (2002) Distinct nuclear organization, DNA methylation pattern and cytokinin distribution mark juvenile, juvenile-like and adult vegetative apical meristems in peach (Prunus persica (L.) Batsch). J Exp Bot 53(371):1047–1054
Cao XF, Jacobsen SE (2002a) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99:16491–16498
Cao X, Jacobsen SE (2002b) Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12(13):1138–1144
Cao X, Springer NM, Muszynski MG, Phillips RL, Kaeppler S, Jacobsen SE (2000) Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc Natl Acad Sci USA 97(9):4979–4984
Cervera MT, Ruiz-García L, Martínez-Zapater J (2002) Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP marker. Mol Genet Genomics 268(4):543–552
Chan SW, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6(5):351–360
Chang L, Zhang Z, Yang H, Li H, Dai H (2007) Detection of strawberry RNA and DNA viruses by RT-PCR using total nucleic acid as a template. J Phytopathol 155:431–436
Dai H, Zhang Z, Guo X (2007) Adventitious bud regeneration from leaf and cotyledon explants of Chinese hawthorn (Crataegus pinnatifida Bge. var. major N.E.Br.). In Vitro Cell Dev Biol Plant 43:2–8
Finnegan EJ, Dennis ES (1993) Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res 21(10):2383–2388
Finnegan EJ, Kovac KA (2000) Plant DNA methyltransferases. Plant Mol Biol 43(2–3):189–201
Finnegan EJ, Peacock WJ, Dennis ES (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93(16):8449–8454
Fraga MF, Cañal MJ, Rodríguez R (2002) Phase-change related epigenetic and physiological changes in Pinus radiata D. Don. Planta 215(4):672–678
Fujimoto R, Sasaki T, Nishio T (2006) Characterization of DNA methyltransferase genes in Brassica rapa. Genes Genet Syst 81(4):235–242
Genger RK, Kovac KA, Dennis ES, Peachock WJ, Finnegan EJ (1999) Multiple DNA methyltransferase genes in Arabidopsis thaliana. Plant Mol Biol 41(2):269–278
Giannino D, Mele G, Cozza R, Bruno L, Testone G, Ticconi C, Frugis G, Bitonti MB, Innocenti AM, Mariotti D (2003) Isolation and characterization of a maintenance DNA-methyltransferase gene from peach (Prunus persica [L.] Batsch): transcript localization in vegetative and reproductive meristems of triple buds. J Exp Bot 54(393):2623–2633
Hasbún R, Valledor L, Berdasco M, Santamaria E, Cañal MJ, Rodriguez R, Rios D, Sánchez M (2005) In vitro proliferation and genome DNA methylation in adult chestnuts. Acta Hort 693:333–340
Henikoff S, Comai L (1998) A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics 149(1):307–318
Jaligot E, Rival A, Beulé T, Dussert S, Vereil JL (2000) Somaclonal variation in oil palm (Elaeis guineensis Jacq.): the DNA methylation hypothesis. Plant Cell Rep 19:684–690
Joyce SM, Cassells AC (2002) Variation in potato microplant morphology in vitro and DNA methylation. Plant Cell Tiss Org Cult 70(2):125–137
Kanai Y, Saito Y, Ushijima S (2004) Alterations in gene expression associated with the overexpression of a splice variant of DNA methyltransferase 3b, DNMT3b4, during human hepatocarcinogenesis. J Cancer Res Clin Oncol 130:636–644
Karhu S (2001) Growth characteristics of micropropagated strawberries. Acta Hort 560:539–542
Kinet JM, Parmentier A (1989) The flowering behaviour of micropropagated strawberry plants cv. ‘Gorella’: the influence of the number of sub-cultures on the multiplication medium. Acta Hort 265:327–334
Kumar S, Cheng X, Klimasauskas S, Mi S, Posfai J, Roberts RJ, Wilson GG (1994) The DNA (cytosine-5) methyltransferases. Nucleic Acids Res 22(1):1–10
Li X, Xu M, Korban SS (2002) DNA methylation profiles differ between field- and in vitro-grown leaves of apple. J Plant Physiol 159:1229–1234
Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of chromomethylase3 for maintenance of CpXpG methylation. Science 292(5524):2077–2080
Litwińczuk W (2004) Field performance of ‘Senga Sengana’ strawberry plants (Fragaria × ananassa Duch.) obtained by runners and in vitro through axillary and adventitious shoots. EJPAU, Horticulture 7(1):art–03
Matthes M, Singh R, Cheah SC, Karp A (2001) Variation in oil palm (Elaeis guineensis Jacq.) tissue culture-derived regenerants revealed by AFLPs with methylation-sensitive enzymes. Theor Appl Genet 102(6–7):971–979
Messeguer R, Ganal MW, Steffens JC, Tanksley SD (1991) Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol Biol 16(5):753–770
Mi J, Zhang Z, Li H, Gao X, Du G (2007) Blossoming and fruiting of micropropagated strawberry plants after transplantation. J Fruit Sci 24(4):472–476
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15:473–497
Nakano Y, Steward N, Sekine M, Kusano T, Sano H (2000) A tobacco NtMET1 cDNA encoding a DNA methyltransferase: molecular characterization and abnormal phenotypes of transgenic tobacco plants. Plant Cell Physiol 41(4):448–457
Papa CM, Springer NM, Muszynski MG, Meeley R, Kaeppler SM (2001) Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell 13:1919–1928
Pósfai J, Bhagwat AS, Pósfait G, Roberts RJ (1989) Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res 17(7):2421–2435
Pradhan S, Cummings M, Roberts RJ, Adams RL (1998) Isolation, characterization and baculovirus-mediated expression of the cDNA encoding cytosine DNA methyltransferase from Pisum sativum. Nucleic Acids Res 26(5):1214–1222
Richards EJ (1997) DNA methylation and plant development. Trends Genet 13:319–323
Rival A, Jaligot E, Beulé T, Finnegan EJ (2008) Isolation and expression analysis of genes encoding MET, CMT, and DRM methyltransferases in oil palm (Elaeis guineensis Jacq.) in relation to the ‘mantled’ somaclonal variation. J Exp Bot 59(12):3271–3281
Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL (1996) Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273(5275):654–657
Simovic N, Wolyn D, Jelenkovic G (1992) Sequence analysis of 18S ribosomal RNA gene in Fragaria × ananassa Duch. cultivated octoploid strawberry. Plant Mol Biol 18(6):1217–1220
Singh A, Zubko E, Meyer P (2008) Cooperative activity of DNA methyltransferases for maintenance of symmetrical and non-symmetrical cytosine methylation in Arabidopsis thaliana. Plant J 56(5):814–823
Steward N, Kusano T, Sano H (2000) Expression of ZmMET1, a gene encoding a DNA methyltransferase from maize, is associated not only with DNA replication in actively proliferating cells, but also with altered DNA methylation status in cold-stressed quiescent cells. Nucleic Acids Res 28(17):3250–3259
Steward N, Ito M, Yamaguchi Y, Koizumi N, Sano H (2002) Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J Biol Chem 277(40):37741–37746
Szczygiel A, Pierzga K, Borkowska B (2002) Performance of micropropagated strawberry plantlets after planting in the field. Acta Hort 567:317–320
Teerawanichpan P, Chandrasekharan MB, Jiang Y, Narangajavana J, Hall TC (2004) Characterization of two rice DNA methyltransferase genes and RNAi-mediated reactivation of a silenced transgene in rice callus. Planta 218(3):337–349
Teyssier E, Bernacchia G, Maury S, How Kit A, Stammitti-Bert L, Rolin D, Gallusci P (2008) Tissue dependent variations of DNA methylation and endoreduplication levels during tomato fruit development and ripening. Planta 228(3):391–399
Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313(5793):1596–1604
Wada Y (2005) Physiological functions of plant DNA methyltransferases. Plant Biotechnol 22(2):71–80
Wada Y, Ohya H, Yamaguchi Y, Koizumi N, Sano H (2003) Preferential de novo methylation of cytosine residues in non-CpG sequences by a domains rearranged DNA methyltransferase from tobacco plants. J Biol Chem 278(43):42386–42393
Xiao M, Zhang Z, Dai H, Yang H, Li H (2005) Enhancing the stability of detection of Strawberry vein banding virus by PCR. J Fruit Sci 22(5):483–487
Xiao W, Custard KD, Brown RC, Lemmon BE, Harada JJ, Goldberg RB, Fischer RL (2006) DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18(4):805–814
Xiong LZ, Xu CG, Saghai Maroof MA, Zhang Q (1999) Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol Gen Genet 261(3):439–446
Zhang X, Zhang Z, Gao X, Li H, Du G (2006) Epigenetic variation in characteristics of the micro-propagated strawberry plants and their offsprings. J Fruit Sci 23(4):542–546
Zhu Y, Zhao HF, Ren GD, Yu XF, Cao SQ, Kuai BK (2005) Characterization of a novel developmentally retarded mutant (drm1) associated with the autonomous flowering pathway in Arabidopsis. Cell Res 15(2):133–140
Zluvova J, Janousek B, Vyskot B (2001) Immunohistochemical study of DNA methylation dynamics during plant development. J Exp Bot 52(365):2265–2273
Acknowledgments
This work was supported by National Natural Science Foundation of China (Grant No. 30671432) and Program for New Century Excellent Talents in University (NCET-07-0565).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Reski.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chang, L., Zhang, Z., Han, B. et al. Isolation of DNA-methyltransferase genes from strawberry (Fragaria × ananassa Duch.) and their expression in relation to micropropagation. Plant Cell Rep 28, 1373–1384 (2009). https://doi.org/10.1007/s00299-009-0737-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-009-0737-8