Abstract
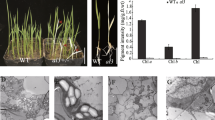
Higher plant chloroplast division involves some of the same types of proteins that are required in prokaryotic cell division. These include two of the three Min proteins, MinD and MinE, encoded by the min operon in bacteria. Noticeably absent from annotated sequences from higher plants is a MinC homologue. A higher plant functional MinC homologue that would interfere with FtsZ polymerization, has yet to be identified. We sought to determine whether expression of the bacterial MinC in higher plants could affect chloroplast division. The Escherichia coli minC (EcMinC) gene was isolated and inserted behind the Arabidopsis thaliana RbcS transit peptide sequence for chloroplast targeting. This TP-EcMinC gene driven by the CaMV 35S2 constitutive promoter was then transformed into tobacco (Nicotiana tabacum L.). Abnormally large chloroplasts were observed in the transgenic plants suggesting that overexpression of the E. coli MinC perturbed higher plant chloroplast division.




Similar content being viewed by others
References
Aldridge C, Maple J, Moller SG (2005) The molecular biology of plastid division in higher plants. J Exp Bot 56:1061–1077
Bramhill D (1997) Bacterial cell division. Annu Rev Cell Dev Biol 13:395–424
Cha JH, Stewart GC (1997) The divIVA minicell locus of Bacillus subtilis. J Bacteriol 179:1671–1683
Colletti KS, Tattersall EA, Pyke KA, Froelich JE, Stokes KD, Osteryoung KW (2000) A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr Biol 10:507–516
de Boer PA, Crossley RE, Rothfield LI (1990) Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci USA 87:1129–1133
de Boer PA, Crossley RE, Rothfield LI (1992) Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J Bacteriol 174:63–70
Dinkins R, Reddy MS, Leng M, Collins GB (2001) Overexpression of the Arabidopsis thaliana MinD1 gene alters chloroplast size and number in transgenic tobacco plants. Planta 214:180–188
Dinkins RD, Conn HM, Dirk LMA, Williams MA, Houtz RL (2003) The Arabidopsis thaliana Peptide Deformylase 1 protein is localized to both mitochondria and chloroplasts. Plant Sci 165:751–758
Emanuelsson O, Nielse H, von Heijne G (1999) ChloroP, a neural network-based method of predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8:978–984
Fu X, Shih YL, Zhang Y, Rothfield LI (2001) The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc Natl Acad Sci USA 98:980–985
Fujiwara MT, Nakamura A, Itoh R, Shimada Y, Yoshida S, Moller SG (2004) Chloroplast division site placement requires dimerization of the ARC11/AtMinD1 protein in Arabidopsis. J Cell Sci 117:2399–2410
Gaikwad A, Babbarwal V, Pant V, Mukherjee SK (2000) Pea chloroplast FtsZ can form multimers and correct the thermosensitive defect of an Escherichia coli ftsZ mutant. Mol Gen Genet 263:213–221
Hale CA, Meinhardt H, de Boer PA (2001) Dynamic localization cycle of the cell division regulator MinE in Escherichia coli. EMBO J 20:1563–1572
Hu Z, Lutkenhaus J (1999) Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol Microbiol 34:82–90
Hu Z, Lutkenhaus J (2000) Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J Bacteriol 182:3965–3971
Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J (1999) The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc Natl Acad Sci USA 96:14819–14824
Itoh R, Fujiwara M, Nagata N, Yoshida S (2001) A chloroplast protein homologous to the eubacterial topological specificity factor minE plays a role in chloroplast division. Plant Physiol 127:1644–1655
Johnson JE, Lackner LL, de Boer PA (2002) Targeting of (D)MinC/MinD and (D)MinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J Bacteriol 184:2951–2962
Kanamaru K, Fujiwara M, Kim M, Nagashima A, Nakazato E, Tanaka K, Takahashi H (2000) Chloroplast targeting, distribution and transcriptional fluctuation of AtMinD1, a Eubacteria-type factor critical for chloroplast division. Plant Cell Physiol 41:1119–1128
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lutkenhaus J (1998) The regulation of bacterial cell division: a time and place for it. Curr Opin Microbiol 1:210–215
Maple J, Aldridge C, Moller SG (2005) Plastid division is mediated by combinatorial assembly of plastid division components. Plant J 43:811–823
Maple J, Chua NH, Moller SG (2002) The topological specificity factor AtMinE1 is essential for correct plastid division site placement in Arabidopsis. Plant J 31:269–277
Margolin W (2001) Bacterial cell division: a moving MinE sweeper boggles the MinD. Curr Biol 11:R395–R398
Marston AL, Errington J (1999) Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol Microbiol 33:84–96
Morrison TB, Weis JJ, Wittwer CT (1998) Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques 24:954–958, 960, 962
Orozco EMJ, Mullet JE, Hanley-Bowdoin L, Chua NH (1986) In vitro transcription of chloroplast protein genes. Methods Enzymol 118:232–253
Osteryoung KW, McAndrew RS (2001) The plastid division machine. Annu Rev Plant Physiol Plant Mol Biol 52:315–333
Osteryoung KW, Nunnari J (2003) The division of endosymbiotic organelles. Science 302:1698–1704
Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY (1998) Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10:1991–2004
Pichoff S, Lutkenhaus J (2001) Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J Bacteriol 183:6630–6635
Ramirez-Arcos S, Greco V, Douglas H, Tessier D, Fan D, Szeto J, Wang J, Dillon JR (2004) Conserved glycines in the C terminus of MinC proteins are implicated in their functionality as cell division inhibitors. J Bacteriol 186:2841–2855
Raskin DM, de Boer PA (1999) MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol 181:6419–6424
Raynaud C, Cassier-Chauvat C, Perennes C, Bergounioux C (2004) An Arabidopsis homolog of the bacterial cell division inhibitor SulA is involved in plastid division. Plant Cell 16:1801–1811
Reddy MS, Dinkins R, Collins GB (2002) Overexpression of the Arabidopsis thaliana MinE1 bacterial division inhibitor homologue gene alters chloroplast size and morphology in transgenic Arabidopsis and tobacco plants. Planta 215:167–176
Ririe KM, Rasmussen RP, Wittwer CT (1997) Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem 245:154–160
Rothfield L (2003) New insights into the developmental history of the bacterial cell division site. J Bacteriol 185:1125–1127
Rothfield L, Justice S, Garcia-Lara J (1999) Bacterial cell division. Annu Rev Genet 33:423–448
Schardl CL, Byrd AD, Benzion G, Altschuler MA, Hildebrand DF, Hunt AG (1987) Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61:1–11
Shih YL, Le T, Rothfield L (2003) Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA 100:7865–7870
Vitha S, Froehlich JE, Koksharova O, Pyke KA, van Erp H, Osteryoung KW (2003) ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15:1918–1933
Vitha S, McAndrew RS, Osteryoung KW (2001) FtsZ ring formation at the chloroplast division site in plants. J Cell Biol 153:111–120
Acknowledgements
The authors thank Dr. Joe Lutkenhaus and Dr. Robert Houtz for antisera to E. coli MinC and the large subunit of ribulose-1,5-bisphosphate carboxylase proteins, respectively; Mellisa Dawdry, Dr. Nihar Nayak and Praveen Pallikonda for technical assistance; Drs. Saratha Kumudini, Archie Portis and Ling Yuan for critical reviews of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Kamo
Rights and permissions
About this article
Cite this article
Tavva, V.S., Collins, G.B. & Dinkins, R.D. Targeted overexpression of the Escherichia coli MinC protein in higher plants results in abnormal chloroplasts. Plant Cell Rep 25, 341–348 (2006). https://doi.org/10.1007/s00299-005-0086-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-005-0086-1