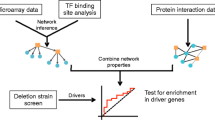
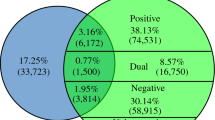
Abstract
Complex biological processes are frequently regulated through networks comprised of multiple signaling pathways, transcription factors, and effector molecules. The identity of specific genes carrying out these functions is usually determined by single mutant genetic analysis. However, to understand how the individual genes/gene products function, it is necessary to determine how they interact with other components of the larger network; one approach to this is to use genetic interaction analysis. The human fungal pathogen Candida albicans regulates biofilm formation through an interconnected set of transcription factor hubs and is, therefore, an example of this type of complex network. Here, we describe experiments and analyses designed to understand how the C. albicans biofilm transcription factor hubs interact and to explore the role of network structure in its overall function. To do so, we analyzed published binding and genetic interaction data to characterize the topology of the network. The hubs are best characterized as a small world network that functions with high efficiency and low robustness (high fragility). Highly efficient networks rapidly transmit perturbations at given nodes to the rest of the network. Consistent with this model, we have found that relatively modest perturbations, such as reduction in the gene dosage of hub transcription factors by one-half, lead to significant alterations in target gene expression and biofilm fitness. C. albicans biofilm formation occurs under very specific environmental conditions and we propose that the fragile, small world structure of the genetic network is part of the mechanism that imposes this stringency.


Similar content being viewed by others
References
Barabási AL, Oltvai ZN (2004) Network biology: understanding the cell’s functional organization. Nat Rev Genet 5:101–113
Bharucha N, Chabrier-Rosello Y, Xu T, Johnson C, Sobczynski S et al (2011) A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM network during morphogenesis. PLoS Genet 7:e1002058
Blank LM, Kuepfer L, Sauer U (2005) Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol 6:R49
Boone C, Bussey H, Andrews BJ (2007) Exploring genetic interactions and networks with yeast. Nat Rev Genet 8:437–449
Du H, Huang G (2016) Environmental pH adaption and morphological transitions in Candida albicans. Curr Genet 62:283–286
Finkel JS, Xu W, Huang D, Hill EM, Desai JV et al (2012) Portrait of Candida albicans adherence regulators. PLoS Pathog 8:e1002525
Fox EP, Bui CK, Nett JE, Hartooni N, Mui MC et al (2015) An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol Microbiol 96:1226–1239
Glazier VE, Murante T, Murante D, Koselny K, Liu Y, Kim D, Koo H, Krysan DJ (2017) Genetic analysis of the Candida albicans biofilm transcription factor network using simple and complex haploinsufficiency. PLoS Genet 13:e1006948
Homann OR, Dea J, Noble SM, Johnson AD (2009) A phenotypic profile of the Candida albicans regulatory network. PLoS Genet 5:e1000783
Köhler JR, Casadevall A, Perfect J (2014) The spectrum of fungi that infects humans. Cold Spring Harb Perspect Med 5:a019273
Kovács ÁT (2016) Bacterial differentiation via gradual activation of global regulators. Curr Genet 62:125–128
Lohse MB, Gulati M, Johnson AD, Nobile CJ (2018) Development and regulation of single- and multi-species Candida albicans biofilms. Nat Rev Microbiol 16:19-3
Masel J, Siegal ML (2009) Robustness: mechanisms and consequences. Trends Genet 25:395–403
McCarty TP, Pappas PG (2016) Invasive candidiasis. Infect Dis Clin North Am 30:103–124
Morine MJ, Gu H, Myers RA, Bielawski JP (2009) Trade-offs between efficiency and robustness in bacterial metabolic networks are associated with niche breadth. J Mol Evol 68:506–515
Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD et al (2012) A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138
Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD (2016) Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin Infect Dis 62:e1–e50
Peng G-S, Tan S-Y, Wu J, Holme P (2016) Trade-offs between robustness and small-world effect in complex networks. Sci Rep 6:37317
Sorrells TR, Johnson AD (2015) Making sense of transcription networks. Cell 161:714–723
Tejima K, Ishiai M, Murayama SO, Iwatani S, Kajiwara S (2017) Candida albicans fatty acyl–CoA synthetase, CaFaa4p, is involved in the uptake of exogenous long-chain fatty acids and cell activity in the biofilm. Curr Genet. https://doi.org/10.1007/s00294-017-0751-2
Watts DJ, Strogatz SH (1998) Collective dynamics of ‘small-world’ networks. Nature 393:440–442
Woolford CA, Lagree K, Aleynikov T, Mitchell AP (2017) Negative control of Candida albicans filamentation-associated gene expression by essential protein kinase gene KIN28. Curr Genet 63:1073–1079
Acknowledgements
This work was supported by NIH Grants F32AI26634 (VEG) and 1R01AI098450 (DJK). We thank Aaron Mitchell (Carnegie Mellon) and Scott Filler (UCLA) for stimulating discussions regarding this project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Rights and permissions
About this article
Cite this article
Glazier, V.E., Krysan, D.J. Transcription factor network efficiency in the regulation of Candida albicans biofilms: it is a small world. Curr Genet 64, 883–888 (2018). https://doi.org/10.1007/s00294-018-0804-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-018-0804-1