Abstract
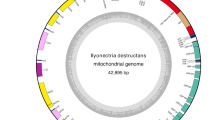
The sequences of the mitochondrial genomes of the oomycetes Phytophthora ramorum and P. sojae were determined during the course of complete nuclear genome sequencing (Tyler et al., Science, 313:1261,2006). Both mitochondrial genomes are circular mapping, with sizes of 39,314 bp for P. ramorum and 42,977 bp for P. sojae. Each contains a total of 37 recognizable protein-encoding genes, 26 or 25 tRNAs (P. ramorum and P. sojae, respectively) specifying 19 amino acids, six more open reading frames (ORFs) that are conserved, presumably due to functional constraint, across Phytophthora species (P. sojae, P. ramorum, and P. infestans), six ORFs that are unique for P. sojae and one that is unique for P. ramorum. Non-coding regions comprise about 11.5 and 18.4% of the genomes of P. ramorum and P. sojae, respectively. Relative to P. sojae, there is an inverted repeat of 1,150 bp in P. ramorum that includes an unassigned unique ORF, a tRNA gene, and adjacent non-coding sequences, but otherwise the gene order in both species is identical. Comparisons of these genomes with published sequences of the P. infestans mitochondrial genome reveals a number of similarities, but the gene order in P. infestans differed in two adjacent locations due to inversions and specific regions of the genomes exhibited greater divergence than others. For example, the breakpoints for the inversions observed in P. infestans corresponded to regions of high sequence divergence in comparisons between P. ramorum and P. sojae and the location of a hypervariable microsatellite sequence (eight repeats of 24 bp) in the P. sojae genome corresponds to a site of major length variation in P. infestans. Although the overwhelming majority of each genome is conserved (81–92%), there are a number of genes that evolve more rapidly than others. Some of these rapidly evolving genes appear specific to Phytophthora, arose recently, and future evaluation of their function and the effects of their loss could prove fruitful for understanding the phylogeny of these devastating plant pathogens.




Similar content being viewed by others
References
Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Gelpke MD, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJ, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S (2002) Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297:1301–1310
Avila-Adame C, Gomez-Alpizar L, Zismann V, Jones KM, Buell CR, Beagle Ristaino J (2006) Mitochondrial genome sequences and molecular evolution of the Irish potato famine pathogen, Phytophthora infestans. Curr Genet 49:39–46
Baldauf SL, Palmer JD (1993) Animals and fungi are each other’s closest relatives: congruent evidence from multiple proteins. Proc Natl Acad Sci USA 90:11558–11562
Bhattacharya D, Stickel SK (1994) Sequence analysis of duplicated actin genes in Lagenidium giganteum and Pythium irregulare (Oomycota). J Mol Evol 39:56–61
Boyd DA, Hobman TC, Gruenke SA, Klassen GR (1984) Evolutionary stability of mitochondrial DNA organization in Achlya. Can J Biochem Cell Biol 62:571–576
Brasier CM, Denman S, Brown A, Webber J (2005) Sudden oak death (Phytophthora ramorum) discovered in trees in Europe. Mycol Res 108:1108–1110
Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S (2003) NISC comparative sequencing program. Lagan and multi-lagan: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 13:721–731
Chesnick JM, Goff M, Graham J, Ocampo C, Lang BF, Seif E, Burger G (2000) The mitochondrial genome of the stramenopile alga Chrysodidymus synuroideus. Complete sequence, gene content and genome organization. Nucleic Acids Res 28:2512–2518
Cooke DEL, Drenth A, Duncan JM, Wagels G, Brasier CM (2000) A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet Biol 30:17–32
Davidson JM, Werres S, Garbelotto M, Hanson E, Rizzo DM (2003) Sudden oak death and associated diseases caused by Phytophthora ramorum. Plant Health Prog. doi:10.1094/PHP-2003-0707-01-DG
Dick MW (2001) Straminipilous fungi. Kluwer, Hingham, MA
Erwin DC, Ribeiro OK (1996) Phytophthora diseases worldwide. American Phytopathological Society Press St. Paul, MN, 562 pp
Förster H, Coffey MD, Elwood H, Sogin ML (1990) Sequence analysis of the small subunit ribosomal RNAs of three zoosporic fungi and implications for fungal evolution. Mycologia 82:306–312
Förster H, Kinscherf TG, Leong S, Maxwell DP (1987) Molecular analysis of the mitochondrial genome of Phytophthora. Curr Genet 12:215–218
Förster H, Kinscherf TG, Leong S, Maxwell DP (1988) Estimation of relatedness between phytophthora species by analysis of mitochondrial DNA. Mycologia 80:466–478
Förster H, Kinscherf TG, Leong S, Maxwell DP (1989) Restriction fragment length polymorphism of the mitochondrial DNA of phytophthora megasperma isolated form soybean, alfalfa, and fruit trees. Can J Bot 67:529–537
Förster H, Tyler BM, Coffey MD (1994) Phytophthora sojae races have arisen by clonal evolution and by rare outcrosses. Mol Plant Microbe Interact 7:780–791
Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32 (web server issue):W273–W279
Gray MW, Lang BF, Cedergren R, Golding G, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn TG, Plante I, Rioux P, Saint-Louis D, Zhu Y, Burger G (1998) Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res 26: 865–878
Grayburn WS, Hudspeth DSS, Gane MK, Hudspeth MES (2004) The mitochondrial genome of Saprolegnia ferax: organization, gene content, and nucleotide sequence. Mycologia 96:980–987
Hudspeth MES, Shumard DS, Bradford JR, Grossman LI (1983) Organization of Achlya mtDNA: a population with two orientation and a large inverted repeat containing the rRNA genes. Proc Natl Acad Sci USA 80:142–146
Knoll HA (1992) The early evolution of eukaryotes: a geological perspective. Science 256:622–627
Kroon LPNM, Bakker FT, van den Bosch GB, Bonnants PJ, Flier WG (2004) Phylogenetic analysis of Phytophthora species based on mitochondrial and nuclear DNA sequences. Fungal Genet Biol 41:766–782
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Martin FN (1991) Linear mitochondrial molecules and intraspecific mitochondrial genome stability in a species of Pythium. Genome 34:156–162
Martin FN (2000) Phylogenetic relationships among some Pythium species inferred from sequence analysis of the mitochondrially encoded cytochrome oxidase II gene. Mycologia 92:711–727
Martin FN, Tooley PW (2003a) Phylogenetic relationships among Phytophthora species inferred from sequence analysis of the mitochondrially-encoded cytochrome oxidase I and II genes. Mycologia 95:269–284
Martin FN, Tooley PW (2003b) Phylogenetic relationships of Phytophthora ramorum, P. nemorosa, and P. pseudosyringae, three species recovered from areas in California with sudden oak death. Mycol Res 107:1379–1391
Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046
McNabb SA, Boyd DA, Belkhiri A, Dick MW, Klassen GR (1987) An inverted repeat comprises more than three quarters of the mitochondrial genome in two species of Pythium. Curr Genet 12:205–208
McNabb SA, Klassen GR (1988) Uniformity of mitochondrial DNA complexity in Oomycetes and the evolution of the inverted repeat. Exp Mycol 12:233–242
McNabb SA, Eros RW, Klassen GR (1988) Presence and absence of large inverted repeats in the mitochondrial DNA of hyphochytriomycetes. Can J Bot 66:2377–2379
Oudot-Le Sec M-P, Loiseaux-de Goër S, Stam WT, Olsen JL (2006) Complete mitochondrial genomes of the three brown algae (heterokonta: phaeophyceae) Dictyota dichotomam, Fucus vesiculosus and Desmarestia viridis. Curr Genet 49:47–58
Paquin B, Laforest M-J, Forget L, Roewer I, Wang Z, Longcore J, Lang BF (1997) The fungal mitochondrial genome project: evolution of fungal mitochondrial genomes and their gene expression. Curr Genet 31:380–395
Rizzo DM, Garbelotto M, Davidson JM, Slaughter GW, Koike ST (2002) Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California. Plant Dis 86:205–214
Shumard-Hudspeth DS, Hudspeth MES (1990) Genetic rearrangements in Phytophthora mitochondrial DNA. Curr Genet 17:413–415
Shumard DS, Grossman LI, Hudspeth MES (1986) Achlya mitochondrial DNA: gene localization and analysis of inverted repeats. Mol Gen Genet 202:16–23
Tyler BM, Tripathi S, Zhang X, Dehal P, Jiang R, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon J, Chapman J, Damasceno CMB, Dorrance AE, Dou D, Dickerman AW, Dubchak I, Garbelotto M, Gijzen M, Gordon SG, Govers F, Grunwald NJ, Huang W, Ivors K, Jones RW, Kamoun S, Krampis K, Lamour K, Lee M-K, McDonald WH, Medina M, Meijer HJG, Nordberg EK, Maclean DJ, Ospina-Giraldo MD, Morris P, Phuntumart V, Putnam N, Rash S, Rose JKC, Sakihama Y, Salamov A, Savidor A, Scheuring CF, Smith BM, Sobral BWS, Terry A, Torto-Alalibo TA, Win J, Xu Z, Zhang H, Grigoriev I, Rokhsar D, Boore JL (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313:1261–1266
Wainright PO, Hinkle G, Sogin ML, Stickel SK (1993) Monophyletic origins of the metazoa: an evolutionary link with fungi. Science 260:340–342
Weerakoon ND, Roberts JK, Lehnen LP, Hardham AR (1998) Isolation and characterization of the single β-tublin gene in Phytophthora cinnamomi. Mycologia 90:85–95
Werres S, Marwitz R, Man In’T Veld WA, Cock AWAM, Bonants PJM, Weerdt Md, Themann K, Ilieva E, Baayen RP (2001) Phytophthora ramorum sp. nov., a new pathogen on Rhododendron and viburnum. Mycol Res 105:1155–1165
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yang Z, Nielsen R (2000) Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 17:32–43
Acknowledgments
Thanks to Susan Lucas and all of the members of the JGI Production Sequencing Department for leading the efforts to determine these sequences. Thanks for related technical support to Jarrod Chapman, Nik Putnam, Dan Rokhsar, Astrid Terry, and Harris Shapiro. This work was supported by National Science Foundation Grant MCB-0242131 and by grant 2002-35600-12747 from the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, and was performed partly under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under Contract No. DE-AC02-05CH11231 and Los Alamos National Laboratory under Contract No. W-7405-ENG-36.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Tomaska.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Martin, F.N., Bensasson, D., Tyler, B.M. et al. Mitochondrial genome sequences and comparative genomics of Phytophthora ramorum and P. sojae . Curr Genet 51, 285–296 (2007). https://doi.org/10.1007/s00294-007-0121-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-007-0121-6