Abstract
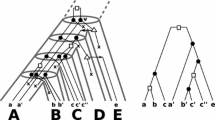
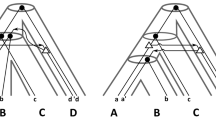
Phylogenomics commonly aims to construct evolutionary trees from genomic sequence information. One way to approach this problem is to first estimate event-labeled gene trees (i.e., rooted trees whose non-leaf vertices are labeled by speciation or gene duplication events), and to then look for a species tree which can be reconciled with this tree through a reconciliation map between the trees. In practice, however, it can happen that there is no such map from a given event-labeled tree to any species tree. An important situation where this might arise is where the species evolution is better represented by a network instead of a tree. In this paper, we therefore consider the problem of reconciling event-labeled trees with species networks. In particular, we prove that any event-labeled gene tree can be reconciled with some network and that, under certain mild assumptions on the gene tree, the network can even be assumed to be multi-arc free. To prove this result, we show that we can always reconcile the gene tree with some multi-labeled (MUL-)tree, which can then be “folded up” to produce the desired reconciliation and network. In addition, we study the interplay between reconciliation maps from event-labeled gene trees to MUL-trees and networks. Our results could be useful for understanding how genomes have evolved after undergoing complex evolutionary events such as polyploidy.








Similar content being viewed by others
References
Altenhoff AM, Dessimoz C (2009) Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput Biol 5:e1000262
Altenhoff AM, Gil M, Gonnet GH, Dessimoz C (2013) Inferring hierarchical orthologous groups from orthologous gene pairs. PLoS One 8(1):e53786
Altenhoff AM et al (2015) The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucl Acids Res 43(D1):D240–D249
Bapteste E, van Iersel L, Janke A, Kelchner S, Kelk S, McInerney JO, Morrison DA, Nakhleh L, Steel M, Stougie L et al (2013) Networks: expanding evolutionary thinking. Trends Genet 29(8):439–441
Chen F, Mackey AJ, Stoeckert CJ, Roos DS (2006) OrthoMCL-db: querying a comprehensive multi-species collection of ortholog groups. Nucl Acids Res 34(S1):D363–D368
Cui Y, Jansson J, Sung WK (2012) Polynomial-time algorithms for building a consensus mul-tree. J Comput Biol 19(9):1073–1088
Czabarka É, Erdős P, Johnson V, Moulton V (2013) Generating functions for multi-labeled trees. Discrete Appl Math 161(1–2):107–117
Dondi R, El-Mabrouk N, Swenson KM (2014) Gene tree correction for reconciliation and species tree inference: complexity and algorithms. J Discrete Algorithms 25:51–65 (23rd Annual Symposium on Combinatorial Pattern Matching)
Dondi R, El-Mabrouk N, Lafond M (2016) Correction of weighted orthology and paralogy relations-complexity and algorithmic results. In: International workshop on algorithms in bioinformatics. Springer, pp 121–136
Dondi R, Lafond M, El-Mabrouk N (2017a) Approximating the correction of weighted and unweighted orthology and paralogy relations. Algorithms Mol Biol 12(1):4
Dondi R, Mauri G, Zoppis I (2017b) Orthology correction for gene tree reconstruction: theoretical and experimental results. Procedia Comput Sci 108:1115–1124 (International Conference on Computational Science, ICCS 2017, 12–14 June 2017, Zurich, Switzerland)
Doyon JP, Chauve C, Hamel S (2009) Space of gene/species trees reconciliations and parsimonious models. J Comput Biol 16(10):1399–1418
Doyon JP, Scornavacca C, Gorbunov KY, Szöllősi GJ, Ranwez V, Berry V (2010) An efficient algorithm for gene/species trees parsimonious reconciliation with losses, duplications and transfers. Springer, Berlin, pp 93–108
Doyon JP, Ranwez V, Daubin V, Berry V (2011) Models, algorithms and programs for phylogeny reconciliation. Brief Bioinform 12(5):392–400
Eulenstein O, Huzurbazar S, Liberles DA (2011) Reconciling phylogenetic trees. In: Evolution after gene duplication, Chap. 10. Wiley, pp 185–206. https://doi.org/10.1002/9780470619902.ch10
Fitch WM (1970) Distinguishing homologous from analogous proteins. Syst Zool 19:99–113
Fitch WM (2000) Homology: a personal view on some of the problems. Trends Genet 16:227–231
Gontier N (2015) Reticulate evolution everywhere. In: Gontier N (ed) Reticulate evolution. Springer, Berlin, pp 1–40
Goodman M, Czelusniak J, Moore GW, Romero-Herrera AE, Matsuda G (1979) Fitting the gene lineage into its species lineage, a parsimony strategy illustrated by cladograms constructed from globin sequences. Syst Biol 28(2):132–163
Gorecki P, Tiuryn J (2006) DLS-trees: a model of evolutionary scenarios. Theor Comput Sci 359(1):378–399
Gregg WT, Ather SH, Hahn MW (2017) Gene-tree reconciliation with mul-trees to resolve polyploidy events. Syst Biol 66(6):1007–1018
Hallett MT, Lagergren J (2001) Efficient algorithms for lateral gene transfer problems. In: Proceedings of the fifth annual international conference on computational biology. ACM, pp 149–156
Hassanzadeh R, Eslahchi C, Sung WK (2014) Do triplets have enough information to construct the multi-labeled phylogenetic tree? PLOS One 9(7):1–10
Hellmuth M (2017) Biologically feasible gene trees, reconciliation maps and informative triples. Algorithms Mol Biol 12(1):23
Hellmuth M, Wieseke N (2016) From sequence data including orthologs, paralogs, and xenologs to gene and species trees. In: Pontarotti P (ed) Evolutionary biology: convergent evolution, evolution of complex traits, concepts and methods. Springer, Cham, pp 373–392
Hellmuth M, Hernandez-Rosales M, Huber KT, Moulton V, Stadler PF, Wieseke N (2013) Orthology relations, symbolic ultrametrics, and cographs. J Math Biol 66(1–2):399–420
Hellmuth M, Stadler PF, Wieseke N (2017) The mathematics of xenology: Di-cographs, symbolic ultrametrics, 2-structures and tree-representable systems of binary relations. J Math Biol 75(1):199–237
Hernandez-Rosales M, Hellmuth M, Wieseke N, Huber KT, Moulton V, Stadler PF (2012) From event-labeled gene trees to species trees. BMC Bioinform 13(Suppl 19):S6
Huber KT, Moulton V (2006) Phylogenetic networks from multi-labelled trees. J Math Biol 52(5):613–632
Huber KT, Oxelman B, Lott M, Moulton V (2006) Reconstructing the evolutionary history of polyploids from multilabeled trees. Mol Biol Evol 23(9):1784–1791
Huber KT, Moulton V, Steel M, Wu T (2016) Folding and unfolding phylogenetic trees and networks. J Math Biol 73(6–7):1761–1780
Huson DH, Scornavacca C (2011) A survey of combinatorial methods for phylogenetic networks. Genome Biol Evol 3:23–35
Huson DH, Rupp R, Scornavacca C (2010) Phylogenetic networks: concepts, algorithms and applications. Cambridge University Press, Cambridge. https://doi.org/10.1017/CBO9780511974076
Lafond M, El-Mabrouk N (2014) Orthology and paralogy constraints: satisfiability and consistency. BMC Genom 15(6):S12
Lafond M, El-Mabrouk N (2015) Orthology relation and gene tree correction: complexity results. In: International workshop on algorithms in bioinformatics. Springer, pp 66–79
Lafond M, Swenson KM, El-Mabrouk N (2012) An optimal reconciliation algorithm for gene trees with polytomies. In: International workshop on algorithms in bioinformatics. Springer, pp 106–122
Lafond M, Dondi R, El-Mabrouk N (2016) The link between orthology relations and gene trees: a correction perspective. Algorithms Mol Biol 11(1):1
Lechner M, Findeiß S, Steiner L, Marz M, Stadler PF, Prohaska SJ (2011) Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinform 12:124
Lechner M, Hernandez-Rosales M, Doerr D, Wiesecke N, Thevenin A, Stoye J, Hartmann RK, Prohaska SJ, Stadler PF (2014) Orthology detection combining clustering and synteny for very large datasets. PLoS ONE 9(8):e105015. https://doi.org/10.1371/journal.pone.0105015
Lott M, Spillner A, Huber KT, Petri A, Oxelman B, Moulton V (2009) Inferring polyploid phylogenies from multiply-labeled gene trees. BMC Evol Biol 9(1):216
Ma B, Li M, Zhang L (2000) From gene trees to species trees. SIAM J Comput 30(3):729–752
Nøjgaard N, Geiß M, Merkle D, Stadler PF, Wieseke N, Hellmuth M (2018) Time-consistent reconciliation maps and forbidden time travel. Algorithms Mol Biol 13(1):2
Page R (1998) Genetree: comparing gene and species phylogenies using reconciled trees. Bioinformatics 14(9):819–820
Posada D (2016) Phylogenomics for systematic biology. Syst Biol 65(3):353–356
Rusin LY, Lyubetskaya E, Gorbunov KY, Lyubetsky V (2014) Reconciliation of gene and species trees. BioMed Res Int 2014:1–22
Scornavacca C, Berry V, Ranwez V (2009) From gene trees to species trees through a supertree approach. In: International conference on language and automata theory and applications. Springer, pp 702–714
Scornavacca C, Mayol JCP, Cardona G (2017) Fast algorithm for the reconciliation of gene trees and LGT networks. J Theor Biol 418:129–137
Sonnhammer E, Östlund G (2015) Inparanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucl Acids Res 43(D1):D234–D239
Steel M (2016) Phylogeny: discrete and random processes in evolution. SIAM, Philadelphia
Stolzer M, Lai H, Xu M, Sathaye D, Vernot B, Durand D (2012) Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 28(18):i409
Szöllősi GJ, Daubin V (2012) Modeling gene family evolution and reconciling phylogenetic discord. In: Anisimova M (ed) Evolutionary genomics: statistical and computational methods, vol 2. Humana Press, Totowa, pp 29–51
Szöllősi GJ, Tannier E, Daubin V, Boussau B (2015) The inference of gene trees with species trees. Syst Biol 64(1):e42
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucl Acids Res 28(1):33–36
To TH, Scornavacca C (2015) Efficient algorithms for reconciling gene trees and species networks via duplication and loss events. BMC Genom 16(10):S6
Tofigh A, Hallett M, Lagergren J (2011) Simultaneous identification of duplications and lateral gene transfers. IEEE/ACM Trans Comput Biol Bioinform 8:517–535
Trachana K, Larsson TA, Powell S, Chen WH, Doerks T, Muller J, Bork P (2011) Orthology prediction methods: a quality assessment using curated protein families. BioEssays 33(10):769–780. https://doi.org/10.1002/bies.201100062
Vernot B, Stolzer M, Goldman A, Durand D (2008) Reconciliation with non-binary species trees. J Comput Biol 15(8):981–1006
Acknowledgements
MH would like to thank the School of Computing Sciences, University of East Anglia, and KH and VM would like to thank the Institute of Mathematics and Computer Science, University of Greifswald, for helping to make two visits possible during which this work was conceived and developed. The authors would also like to thank the anonymous referees for their helpful comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hellmuth, M., Huber, K.T. & Moulton, V. Reconciling event-labeled gene trees with MUL-trees and species networks. J. Math. Biol. 79, 1885–1925 (2019). https://doi.org/10.1007/s00285-019-01414-8
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-019-01414-8
Keywords
- Tree reconciliation
- Network reconciliation
- Gene evolution
- Species evolution
- Phylogenetic network
- MUL tree
- Triples