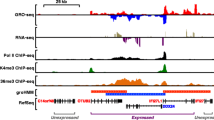
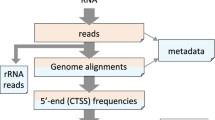
Abstract
A mixture model and statistical method is proposed to interpret the distribution of reads from a nascent transcriptional assay, such as global run-on sequencing (GRO-seq) data. The model is annotation agnostic and leverages on current understanding of the behavior of RNA polymerase II. Briefly, it assumes that polymerase loads at key positions (transcription start sites) within the genome. Once loaded, polymerase either remains in the initiation form (with some probability) or transitions into an elongating form (with the remaining probability). The model can be fit genome-wide, allowing patterns of Pol II behavior to be assessed on each distinct transcript. Furthermore, it allows for the first time a principled approach to distinguishing the initiation signal from the elongation signal; in particular, it implies a data driven method for calculating the pausing index, a commonly used metric that informs on the behavior of RNA polymerase II. We demonstrate that this approach improves on existing analyses of GRO-seq data and uncovers a novel biological understanding of the impact of knocking down the Male Specific Lethal (MSL) complex in Drosophilia melanogaster.







Similar content being viewed by others
References
Adelman K, Lis JT (2012) Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13(10):720–731. doi:10.1038/nrg3293
Allison KA, Kaikkonen MU, Gaasterland T, Glass CK (2013) Vespucci: a system for building annotated databases of nascent transcripts. Nucleic Acids Research. doi:10.1093/nar/gkt1237. http://nar.oxfordjournals.org/content/early/2013/12/04/nar.gkt1237.abstract
Azofeifa J, Allen MA, Lladser ME, Dowell R (2014) FStitch: A fast and simple algorithm for detecting nascent RNA transcripts. In: Proceedings of the 5th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics, BCB ’14, pp. 174–183. ACM, New York, NY, USA. doi:10.1145/2649387.2649427
Azofeifa J, Allen MA, Lladser ME, Dowell R (2016) An annotation agnostic algorithm for detecting nascent RNA transcripts in GRO-seq. IEEE/ACM Trans Comput Biol Bioinform. doi:10.1109/TCBB.2016.2520919
Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, Bentley D (2012) mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Molecular Cell 46(3): 311 – 324 doi:10.1016/j.molcel.2012.03.006. http://www.sciencedirect.com/science/article/pii/S1097276512002134
Core L, Lis J (2008) Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319:1791. doi:10.1126/science.1150843
Danko CG, Hyland SL, Core LJ, Martins AL, Waters CT, Lee HW, Cheung VG, Kraus WL, Lis JT, Siepel A (2014) Accurate identification of active transcriptional regulatory elements from global run-on and sequencing data. bioRxiv. doi:10.1101/011353
Dempster A, Laird N, Rubin D (1977) Maximum likelihood from incomplete data via the EM algorithm. J Royal Stat Soc 39(1):1–38
Durrett R (1999) Essentials of stochastic processes. Springer texts in statistics, 1st edn. Springer, New York. ISBN-13: 978-0387988368, ISBN-10: 038798836X
Fuda NJ, Ardehali MB, Lis JT (2009) Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461(7261):186–192. doi:10.1038/nature08449
Gilchrist DA, Nechaev S, Lee C, Ghosh SKB, Collins JB, Li L, Gilmour DS, Adelman K (2008) Nelf-mediated stalling of pol ii can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes & Development 22(14):1921–1933. doi:10.1101/gad.1643208. http://genesdev.cshlp.org/content/22/14/1921.abstract
Hah N, Danko C, Core L, Waterfall J, Siepel A, Lis J, Kraus W (2011) A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145(4):622–634. doi:10.1016/j.cell.2011.03.042 http://www.sciencedirect.com/science/article/pii/S009286741100376X
Jonkers I, Kwak H, Lis JT (2014) Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 3 doi:10.7554/eLife.02407
Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI (2011) X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature 471(7336):115–118. doi:10.1038/nature09757
Lucchesi JC (1998) Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Current Opinion in Genetics & Development 8(2):179–184. doi:10.1016/S0959-437X(98)80139-1. www.sciencedirect.com/science/article/pii/S0959437X98801391
Melgar M, Collins F, Sethupathy P (2011) Discovery of active enhancers through bidirectional expression of short transcripts. Genome Biology 12(11):R113. doi:10.1186/gb-2011-12-11-r113. http://genomebiology.com/2011/12/11/R113
Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT (2011) Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes & Development 25(7):742–754. doi:10.1101/gad.2005511. http://genesdev.cshlp.org/content/25/7/742.abstract
Paredes SH, Melgar MF, Sethupathy P (2013) Promoter-proximal CCCTC-factor binding is associated with an increase in the transcriptional pausing index. Bioinformatics 29(12):1485–1487 doi:10.1093/bioinformatics/bts596. http://bioinformatics.oxfordjournals.org/content/29/12/1485.abstract
Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA (2010) c-Myc regulates transcriptional pause release. Cell 141(3):432–445. doi:10.1016/j.cell.2010.03.030 http://www.sciencedirect.com/science/article/pii/S0092867410003181
Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA (2008) Divergent transcription from active promoters. Science 322(5909):1849–1851. doi:10.1126/science.1162253 http://www.sciencemag.org/content/322/5909/1849.abstract
Sims RJ, Belotserkovskaya R, Reinberg D (2004) Elongation by RNA polymerase II: the short and long of it. Genes and Development 18(20):2437–2468. doi:10.1101/gad.1235904 http://genesdev.cshlp.org/content/18/20/2437.abstract
Williams LH, Fromm G, Gokey NG, Henriques T, Muse GW, Burkholder A, Fargo DC, Hu G, Adelman K (2015) Pausing of RNA Polymerase II regulates mammalian developmental potential through control of signaling networks. Molecular Cell 58(2):311–322. doi:10.1016/j.molcel.2015.02.003
Acknowledgments
We would like to thank Josephina Hendrix for assistance with analysis of publicly available datasets. This work was funded in part by a NSF IGERT Grant number 1144807 (MEL, JGA, RDD), a Sie Postdoctoral Fellowship (MAA), the Boettcher Foundation’s Webb-Waring Biomedical Research program (RDD) and a NSF ABI DBI-12624L0 (RDD). The authors acknowledge the BioFrontiers Computing Core at the University of Colorado Boulder for providing High Performance Computing resources (NIH 1S10OD012300) supported by BioFrontiers’ IT.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No competing financial interests exist.
Appendix: Double geometric distribution
Appendix: Double geometric distribution
A random variable X is said to have a (possibly asymmetric) Double Geometric distribution with parameters (u, d) when it has the same distribution as \((-U)+D\), where U and D are independent Geometric random variables with means \((1/u-1)\) and \((1/d-1)\), respectively. In particular, the probability mass function of X is
In this case, we write \(X\sim DoubleGeometric(u,d)\). More generally, given an integer i, we write \(X\sim DoubleGeometric(i,u,d)\) to mean that \((X-i)\sim DoubleGeometric(u,d)\).
If \(X\sim DoubleGeometric(i,u,d)\) then
-
(1)
\((i-X)\sim Geometric(u)\) when \(X\le i\); and
-
(2)
\((X-i)\sim Geometric(d)\) when \(X\ge i\).
These two properties justify the Double Geometric terminology.
Rights and permissions
About this article
Cite this article
Lladser, M.E., Azofeifa, J.G., Allen, M.A. et al. RNA Pol II transcription model and interpretation of GRO-seq data. J. Math. Biol. 74, 77–97 (2017). https://doi.org/10.1007/s00285-016-1014-4
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-016-1014-4