Abstract
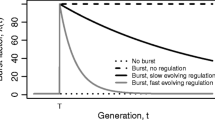
Since its discovery, mobile DNA has fascinated researchers. In particular, many researchers have debated why insertion sequences persist in prokaryote genomes and populations. While some authors think that insertion sequences persist only because of occasional beneficial effects they have on their hosts, others argue that horizontal gene transfer is strong enough to overcome their generally detrimental effects. In this study, we model the long-term fate of a prokaryote cell population, of which a small proportion of cells has been infected with one insertion sequence per cell. Based on our model and the distribution of IS5, an insertion sequence for which sufficient data is available in 525 fully sequenced proteobacterial genomes, we show that the fitness cost of insertion sequences is so small that they are effectively neutral or only slightly detrimental. We also show that an insertion sequence infection can persist and reach the empirically observed distribution if the rate of horizontal gene transfer is at least as large as the fitness cost, and that this rate is well within the rates of horizontal gene transfer observed in nature. In addition, we show that the time needed to reach the observed prevalence of IS5 is unrealistically long for the fitness cost and horizontal gene transfer rate that we computed. Occasional beneficial effects may thus have played an important role in the fast spreading of insertion sequences like IS5.
Similar content being viewed by others
References
Berg DE (1989) Transposon Tn5. In: Berg DE, Howe MM (eds) Mobile DNA. American Society for Microbiology, Washington, D.C., pp 185–210
Bichsel M, Barbour AD, Wagner A (2010) The early phase of a bacterial insertion sequence infection. Theor Popul Biol 78: 278–288
Blot M (1994) Transposable elements and adaptation of host bacteria. Genetica 93(1-3): 5–12
Chandler M, Mahillon J (2002) Insertion sequences revisited. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. American Society for Microbiology, Washington, D.C., pp 305–366
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371: 215–219
Dahlberg C, Bergström M, Hermansson M (1998) In situ detection of high levels of horizontal plasmid transfer in marine bacterial communities. Appl Environ Microbiol 64(7): 2670–2675
Dawkins R (1976) The selfish gene. Oxford University Press, Oxford
Doolittle WF, Sapienza C (1980) Selfish genes, the phenotype paradigm and genome evolution. Nature 284: 601–603
Dröge M, Pühler A, Selbitschka W (1999) Horizontal gene transfer among bacteria in terrestrial and aquatic habitats as assessed by microcosm and field studies. Biol Fertil Soils 29(3): 221–245
Efron B, Tibshirani RJ (1994) An introduction to the bootstrap. Chapman & Hall/CRC, New York
Galas DJ, Chandler M (1989) Bacterial insertion sequences. In: Berg DE, Howe MM (eds) Mobile DNA. American Society for Microbiology, Washington, D.C., pp 109–162
Gibbons RJ, Kapsimalis B (1967) Estimates of the overall rate of growth of the intestinal microflora of hamsters, guinea pigs, and mice. J Bacteriol 93(1): 510–512
Hall BG (1999) Transposable elements as activators of cryptic genes in E. coli. Genetica 107: 181–187
Jiang SC, Paul JH (1998) Gene transfer by transduction in the marine environment. Appl Environ Microbiol 64(8): 2780–2787
Kleckner N (1989) Transposon Tn10. In: Berg DE, Howe MM (eds) Mobile DNA. American Society for Microbiology, Washington, D.C., pp 227–268
Lan R, Reeves PR (2002) Escherichia coli in disguise: molecular origins of shigella. Microbes Infect 4(11): 1125–1132
Lynch M (2007) The origins of genome architecture. Sinauer Associates, Inc, Sunderland
Madigan MT, Martinko JM, Dunlap PV, Clark DP (2009) Brock biology of microorganisms, 12th edn. Pearson Benjamin Cummings, San Francisco
Mahillon J, Siguier P, Chandler M (2009) IS Finder. http://www-is.biotoul.fr
NCBI (2011) National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov
Nelder JA, Mead R (1965) A simplex-method for function minimization. Comput J 7(4): 308–313
Nuzhdin SV (1999) Sure facts, speculations, and open questions about the evolution of transposable element copy number. Genetica 107: 129–137
Orgel LE, Crick FHC (1980) Selfish DNA: the ultimate parasite. Nature 284: 604–607
Savageau MA (1983) Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am Naturalist 122(6): 732–744
Sawyer SA, Dykhuizen DE, DuBose RF, Green L, Mutangadura-Mhlanga T, Wolczyk DF, Hartl DL (1987) Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics 115: 51–63
Schneider D, Lenski RE (2004) Dynamics of insertion sequence elements during experimental evolution of bacteria. Res Microbiol 155: 319–327
Seneta E (1981) Non-negative matrices and Markov chains. Springer, New York
Shapiro JA (1999) Transposable elements as the key to a 21st century view of evolution. Genetica 107: 171–179
So M, McCarthy BJ (1980) Nucleotide sequence of the bacterial transposon TN1681 encoding a heat-stable (ST) toxin and its identification in enterotoxigenic Escherichia coli strains. Proc Natl Acad Sci USA 77(7): 4011–4015
Tavakoli NP, Derbyshire KM (2001) Tipping the balance between replicative and simple transposition. EMBO J 20(11): 2923–2930
Top EM, Springael D (2003) The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol 14: 262–269
Touchon M, Rocha EPC (2007) Causes of insertion sequences abundance in prokaryotic genomes. Mol Biol Evol 24(4): 969–981
Wagner A (2006) Periodic extinctions of transposable elements in bacterial lineages: evidence from intragenomic variation in multiple genomes. Mol Biol Evol 23(4): 723–733
Wagner A, Lewis C, Bichsel M (2007) A survey of bacterial insertion sequences using IScan. Nucleic Acids Res 35(16): 5284–5293
Williams HG, Day MJ, Fry JC, Stewart GJ (1996) Natural transformation in river epilithon. Appl Environ Microbiol 62(8): 2994–2998
Wolfram S (2003) The mathematica book, 5th edn. Wolfram Media, Champaign
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bichsel, M., Barbour, A.D. & Wagner, A. Estimating the fitness effect of an insertion sequence. J. Math. Biol. 66, 95–114 (2013). https://doi.org/10.1007/s00285-012-0504-2
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-012-0504-2
Keywords
- Mobile DNA
- Insertion sequence
- Fitness effect
- Horizontal gene transfer
- Ordinary differential equation system
- Maximum likelihood