Abstract
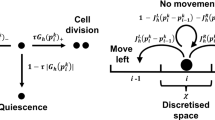
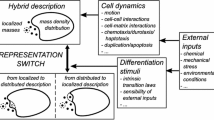
In this paper we compare two alternative theoretical approaches for simulating the growth of cell aggregates in vitro: individual cell (agent)-based models and continuum models. We show by a quantitative analysis of both a biophysical agent-based and a continuum mechanical model that for densely packed aggregates the expansion of the cell population is dominated by cell proliferation controlled by mechanical stress. The biophysical agent-based model introduced earlier (Drasdo and Hoehme in Phys Biol 2:133–147, 2005) approximates each cell as an isotropic, homogeneous, elastic, spherical object parameterised by measurable biophysical and cell-biological quantities and has been shown by comparison to experimental findings to explain the growth patterns of dense monolayers and multicellular spheroids. Both models exhibit the same growth kinetics, with initial exponential growth of the population size and aggregate diameter followed by linear growth of the diameter and power-law growth of the cell population size. Very sparse monolayers can be explained by a very small or absent cell–cell adhesion and large random cell migration. In this case the expansion speed is not controlled by mechanical stress but by random cell migration and can be modelled by the Fisher–Kolmogorov–Petrovskii–Piskounov (FKPP) reaction–diffusion equation. The growth kinetics differs from that of densely packed aggregates in that the initial spread, as quantified by the radius of gyration, is diffusive. Since simulations of the lattice-free agent-based model in the case of very large random migration are too long to be practical, lattice-based cellular automaton (CA) models have to be used for a quantitative analysis of sparse monolayers. Analysis of these dense monolayers leads to the identification of a critical parameter of the CA model so that eventually a hierarchy of three model types (a detailed biophysical lattice-free model, a rule-based cellular automaton and a continuum approach) emerge which yield the same growth pattern for dense and sparse cell aggregates.
Similar content being viewed by others
References
Adam J, Belomo N (1997) A survey of models for tumor-immune system dynamics. Birkhäuser, Boston
Alarcon T, Byrne H, Maini P (2004) A mathematical model of the effects of hypoxia on the cell-cycle of normal and cancer cells. J Theor Biol 229: 395–411
Alber MS, Kiskowski MA, Glazier JA, Jiang Y (2002) On cellular automaton approaches to modeling biological cells. In: Rosenthal J, Gilliam DS (eds) Mathematical systems theory in biology, communication, and finance, IMA 142. Springer, New York, pp 1–40
Alcaraz J, Buscemi L, Grabulosa M, Trepat X, Fabry B, Farre R, Navajas D (2003) Microrheology of human lung epithelial cells measured by atomic force microscopy. Biophys J 84: 2071–2079
Allen M, Tildersley D (1987) Computer Simulation of Liquids. Oxford Science Publications, Oxford
Ambrosi D, Mollica F (2002) On the mechanics of a growing tumor. Int J Eng Sci 40(12): 1297–1316
Ambrosi D, Mollica F (2004) The role of stress in the growth of a multicell spheroid. J Math Biol 48(5): 477–479
Anderson A, Chaplain MAJ, Rejniak K (2007) Single-cell-based models in biology and medicine. Birkhäuser, Basel
Anderson A, Chaplain MAJ, Newman E, Steele R, Thompson A (2000) Mathematical modeling of tumor invasion and metastasis. J Theor Med 2: 129–154
Araujo R, McElwain D (2004) A history of the study of solid tumour growth: the contribution of mathematical models. Bull Math Biol 66: 1039–1091
Beysens D, Forgacs G, Glazier J (2000) Cell sorting is analogous to phase ordering in fluids. Proc Natl Acad Sci USA 97(17): 9467–9471
Block M, Schoell E, Drasdo D (2007) Classifying the expansion kinetics and critical surface dynamics of growing cell populations. Phys Rev Lett 99: 248,101–248,104
Breward C, Byrne H, Lewis C (2002) The role of cell–cell interactions in a two-phase model for avasular tumour growth. J Math Biol 45: 125–152
Bru A, Albertos S, Subiza J, Garcia-Arsenio J, Bru I (2003) The universal dynamics of tumor growth. Biophys J 85: 2948–2961
Bru A, Pastor J, Fernaud I, Bru I, Melle S, Berenguer C (1998) Super-rough dynamics of tumor growth. Phys Rev Lett 81(18): 4008–4011
Byrne H (1997) The importance of intercellular adhesion in the development of carcinomas. IMA J Math Appl Med Biol 14: 305–323
Byrne H, Chaplain J (1996) Modelling the role of cell–cell adhesion in the growth and development of carcinomas. Math Comput Model 12: 1–17
Byrne H, Chaplain M (1997) Free boundary value problem associated with the growth and development of multicellular spheroids. Eur J Appl Math 8: 639–658
Byrne HM, King JR, McElwain DLS, Preziosi L (2003) A two-phase model of solid tumor growth. Appl Math Lett 16(4): 567–573
Byrne H, Preziosi L (2003) Modelling solid tumour growth using the theory of mixtures. Math Med Biol 20: 341–366
Carpick R, Ogletree DF, Salmeron M (1999) A gerneral equation for fitting contact area and friction vs load measurements. J Colloid Interface Sci 211: 395–400
Chaplain M, Graziano L, Preziosi L (2006) Mathematical modelling of the loss of of tissue compression responsiveness and its role in solid tumour development. Math Med Biol 23(3): 197–229
Chen C, Byrne H, King J (2001) The influence of growth-induced stress from the surrounding medium on the development of multicell spheroids. J Math Biol 43: 191–220
Chesla S, Selvaraj P, Zhu C (1998) Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys J 75: 1553–1557
Chu YS et al (2005) Johnson–Kendall–Roberts theory applied to living cells. Phys Rev Lett 94: 028,102
Cickovski T, Huang C, Chaturvedi R, Glimm T, Hentschel H, Alber M, Glazier JA, Newman SA, Izaguirre JA (2005) A framework for three-dimensional simulation of morphogenesis. IEEE/ACM Trans Comput Biol Bioinformatics 2(3): 273–288
Please CP, Pettet G, McElwain D (1998) A new approach to modelling the formation of necrotic regions in tumours. Appl Math Lett 11: 89–94
Cristini V, Lowengrub J, Nie Q (2003) Nonlinear simulations of tumor growth. J Math Biol 46: 191–224
Dallon J, Othmer H (2004) How cellular movement determines the collective force generated by the dictyostelium discoideum slug. J Theor Biol 231: 203–222
DeMasi A, Luckhaus S, Presutti E (2005) Two-scale hydrodynamic limit for a model of malignant tumor cells. MPI-MIS (Preprint) 2: 1–47
Dormann S, Deutsch A (2002) Modeling of self-organized avascular tumor growth with a hybrid cellular automaton. In Silico Biol 2: 0035
Drasdo D (1996) Different growth regimes found in a monte-carlo model of growing tissue cell populations. In: Schweitzer F (eds) Self organization of complex structures: from individual to collective dynamics. Gordon & Breach, New York, pp 281–291
Drasdo D (2003) On selected individual-based approaches to the dynamics of multicellular systems. In: Alt W, Chaplain M, Griebel M (eds) Multiscale modeling. Birkhäuser, Basel
Drasdo D (2005) Coarse graining in simulated cell populations. Adv Complex Syst 8(2 & 3): 319–363
Drasdo D (2008) Center-based single-cell models: an approach to multi-cellular organization based on a conceptual analogy to colloidal particles. In: Anderson A, Chaplain M, Rejniak K (eds) Single-cell-based models in biology and medicine. Birkhäuser, Basel (in press)
Drasdo D, Forgacs G (2000) Modelling the interplay of generic and genetic mechanisms in cleavage, blastulation and gastrulation. Dev Dyn 219: 182–191
Drasdo D, Hoehme S (2005) A single-cell based model to tumor growth in-vitro: monolayers and spheroids. Phys Biol 2: 133–147
Drasdo D, Hoehme S, Block M (2007) On the role of physics in the growth and pattern formation of multi-cellular systems: What can we learn from individual-cell based models?. J Stat Phys 128(1 & 2): 319–363
Drasdo D, Höhme S (2003) Individual-based approaches to birth and death in avascular tumors. Math Comput Model 37: 1163–1175
Drasdo D, Kree R, McCaskill J (1995) Monte-carlo approach to tissue-cell populations. Phys Rev E 52(6): 6635–6657
Friedman A (2007) Mathematical analysis and challenges arrising from models of tumor growth. Math Model Methods Appl Sci 17: 1751–1772
Galle J, Aust G, Schaller G, Beyer T, Drasdo D (2006) Single-cell based mathematical models to the spatio-temporal pattern formation in multi-cellular systems. Cytometry A (in press)
Galle J, Loeffler M, Drasdo D (2005) Modelling the effect of deregulated proliferation and apoptosis on the growth dynamics of epithelial cell populations in vitro. Biophys J 88: 62–75
Galle J, Sittig D, Hanisch I, Wobus M, Wandel E, Loeffler M, Aust G (2006) Individual cell-based models of tumor-environment interactions: multiple effects of cd97 on tumor invasion. J Am Path 169(5): 1802–1811
Graner F, Glazier J (1992) Simulation of biological cell sorting using a two-dimensional extended potts model. Phys Rev Lett 69(13): 2013–2016
Greenspan H (1976) On the growth and stability of cell cultures and solid tumors. J Theor Biol 56(1): 229–242
Hogeweg P (2000) Evolving mechanisms of morphogenesis: on the interplay between differential adhesion and cell differentiation. J Theor Biol 203: 317–333
Johnson K, Kendall K, Roberts A (1971) Surface energy and the contact of elastic solids. Proc R Soc A 324: 301–313
King J, Franks S (2004) Mathematical analysis of some multidimensional tissue-growth models. Eur J Appl Math 15(3): 273–295
Landman K, Please C (2001) The influence of growth-induced stress from the surrounding medium on the development of multicell spheroids. J Math Biol 43: 191–220
Preziosi L (2003) Cancer modelling and simulation. Chapman & Hall/CRC Press, London/West Palm Beach
MacArthur B, Please C (2004) Residual stress generation and necrosis formation in multicell tumour spheroids. J Math Biol 49: 537–552
Mahaffy R, Shih C, McKintosh F, Kaes J (2000) Scanning probe-based frequency-dependent microrheology of polymer gels and biological cells. Phys Rev Lett 85: 880–883
Merks R, Glazier J (2005) A cell-centered approach to developmental biology. Physica A 352: 113–130
Metropolis N, Rosenbluth A, Rosenbluth M, Teller A, Teller E (1953) Equation of state calculations by fast computing machines. J Chem Phys 21: 1087–1092
Lekka M, Laidler P, Gil D, Lekki J, Stachura Z, Hrynkiewicz AZ (1999) Elasticity of normal and cancerous human bladder cells studied by scanning force microscopy. Eur Biophys J 28(4): 312–316
Mombach J, Glazier J (1996) Single cell motion in aggregates of embryonic cells. Phys Rev Lett 76(16): 3032–3035
Moreira J, Deutsch A (2002) Cellular automata models of tumour development—a critical review. Adv Complex Syst 5(1): 247–267
Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS (2005) Proc Natl Acad Sci USA 102: 11594–11599
Newman T (2005) Modeling multi-cellular systems using sub-cellular elements. Math Biosci Eng 2(3): 613–624
Palsson E, Othmer H (2000) A model for individual and collective cell movement in dictyostelium discoideum. Proc Natl Acad Sci USA 12(18): 10,448–10,453
Piper J, Swerlick R, Zhu C (1998) Determining force dependence of two-dimensional receptor-ligand binding affinity by centrifugation. Biophys J 74: 492–513
Preziosi L, Tosin A, Multiphase modeling of tumor growth and extracellular matrix interaction: mathematical tools and applications. J Math Biol
Ramis-Conde I, Chaplain MAJ, Anderson A (2008) Mathematical modelling of cancer cell invasion of tissue. Math Comput Model (in press)
Roose T, Chapman S, Maini P (2007) Mathematical models of avascular tumour growth: a review. SIAM Rev 49(2): 179–208
Roose T, Netti PA, Munn L, Boucher Y, Jain R (2003) Solid stress generated by spheroid growth estimated using a linear poroelasticity model. Microvasc Res 66: 204–212
Schaller G, Meyer-Hermann M (2005) Multicellular tumor spheroid in an off-lattice voronoi-delaunay cell model. Phys Rev E 71: 051,910-1–051,910-16
Schienbein M, Franke K, Gruler H (1994) Random walk and directed movement: comparison between inert particles and self-organized molecular machines. Phys Rev E 49(6): 5462–5471
Schiffer I, Gebhard S, Heimerdinger C, Heling A, Hast J, Wollscheid U, Seliger B, Tanner B, Gilbert S, Beckers T, Baasner S, Brenner W, Spangenberg C, Prawitt D, Trost T, Schreiber W, Zabel B, Thelen M, Lehr H, Oesch F, Hengstler J (2003) Switching off her-2/neu in a tetracycline-controlled mouse tumor model leads to apoptosis and tumor-size-dependent remission. Cancer Res 63: 7221–7231
Stevens A (2000) The derivation of chemotaxis equations as limit dynamics of moderately interacting stochastic many-particle systems. SIAM J Appl Math 61(1): 183–212
Stott E, Britton N, Glazier J, Zajac M (1999) Stochastic simulation of benign avascular tumor growth using the potts model. Math Comput Model 30: 183–198
Ward J, King J (1997) Mathematical modelling of avascular-tumor growth. IMA J Math Appl Med Biol 14: 39–69
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Byrne, H., Drasdo, D. Individual-based and continuum models of growing cell populations: a comparison. J. Math. Biol. 58, 657–687 (2009). https://doi.org/10.1007/s00285-008-0212-0
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-008-0212-0