Abstract
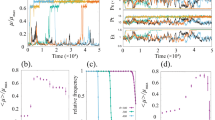
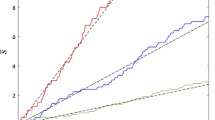
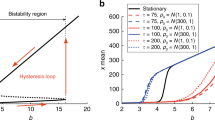
Organisms are known to adapt to regularly varying environments. However, in most cases, the fluctuations of the environment are irregular and stochastic, alternating between favorable and unfavorable regimes, so that cells must cope with an uncertain future. A possible response is population diversification. We assume here that the cell population is divided into two groups, corresponding to two phenotypes, having distinct growth rates, and that cells can switch randomly their phenotypes. In static environments, the net growth rate is maximized when the population is homogeneously composed of cells having the largest growth rate. In random environments, growth rates fluctuate and observations reveal that sometimes heterogeneous populations have a larger net growth rate than homogeneous ones, a fact illustrated recently through Monte-Carlo simulations based on a birth and migration process in a random environment. We study this process mathematically by focusing on the proportion f(t) of cells having the largest growth rate at time t, and give explicitly the related steady state distribution π. We also prove the convergence of empirical averages along trajectories to the first moment \({\mathbb{E}_\pi(f)}\) , and provide efficient numerical methods for computing \({\mathbb{E}_\pi(f)}\) .
Similar content being viewed by others
References
Arkin A., Ross J. and McAdams H. (1998). Stochastic kinetics analysis of developmental pathway bifurcation in phage λ-infected Escherichia coli cells. Genetics 149: 1633–1648
Balaban N., Merrin J., Chait R., Kowalik L. and Leibler S. (2004). Bacterial persistence as a phenotypic switch. Science 305: 1622–1625
Biggar S. and Crabtree G. (2001). Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 20: 3167–3176
Blake W., Kaern M., Cantor C. and Collins J. (2003). Noise in eukaryotic gene expression. Nature 422: 633–637
Breiman L. (1960). The strong law of large numbers for a class of Markov chains. Ann. Math. Stat. 31: 801–803
Bürger R. (1999). Evolution of genetic variability and the advantage of sex and recombination in changing environments. Genetics 153: 1055–1069
Diaconis P. and Freedman D. (1999). Iterated random functions. Siam Rev. 41: 45–76
Dubnau D. and Losick R. (2006). Bistability in bacteria. Molec. Microbio. 61: 564–572
Erdelyi, A.: Higher transcendental functions. Bateman Manuscript Project. vol.1. McGraw-Hill (1953)
Ferrell J. (2002). Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14(2): 140–148
Graumann P. (2006). Different genetic programs within identical bacteria under identical conditions: the phenomenon of bistability greatly modifies our view on bacterial populations. Molec. Microbiol. 61: 560–563
Hairer H., Norsett S. and Wanner G. (2000). Solving ordinary differential equations I, nonstiff problems, Springer series in computational mathematics. Springer, Heidelberg
Jänich K. (2001). Analysis für Physiker und Ingenieure. Springer, Heidelberg
Kearns D. and Losick R. (2005). Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19: 3083–3094
Keren I., Shah D., Spoering A., Kaldalu N. and Lewis K. (2004). Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186: 8172–8180
Kussell E. and Leibler S. (2005). Phenotypic diversity, population growth and information in fluctuating environments. Science 309: 2075–2078
Kussell E., Kishony R., Balaban N. and Leibler S. (2005). Bacterial persistence: a model of survival in changing environments. Genetics 169: 1807–1814
Smits W., Kuipers O. and Veening J. (2006). Phenotypic variation in bacteria: the role of feedback regulation. Nat. Rev. Microbiol. 4: 259–271
Tanaka M., Bergstrom C. and Levin B. (2003). The evolution of mutator genes in bacterial populations: the roles of environmental change and timing. Genetics 164: 843–854
Thattai M. and van Oudenaarden A. (2004). Stochastic gene expression in fluctuating environments. Genetics 167: 523–530
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gander, M.J., Mazza, C. & Rummler, H. Stochastic gene expression in switching environments. J. Math. Biol. 55, 249–269 (2007). https://doi.org/10.1007/s00285-007-0083-9
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-007-0083-9