Abstract
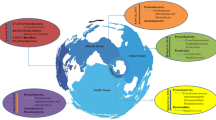
Symbiotic marine bacteria have a pivotal role in drug discovery due to the synthesis of diverse biologically potential compounds. The marine bacterial phyla proteobacteria, actinobacteria and firmicutes are commonly associated with marine macro organisms and frequently reported as dominant bioactive compound producers. They can produce biologically active compounds that possess antimicrobial, antiviral, antitumor, antibiofilm and antifouling properties. Synthesis of these bioactive compounds is controlled by a set of genes of their genomes that is known as biosynthesis gene clusters (BGCs). The development in the field of biotechnology and bioinformatics has uncovered the potential BGCs of the bacterial genome and its functions. Now-a-days researchers have focused their attention on the identification of potential BGCs for the discovery of novel bioactive compounds using advanced technology. This review highlights the marine bacterial symbionts and their BGCs which are responsible for the synthesis of bioactive compounds.



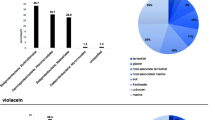
source platform Antismash (bacterial version 5.0). a Shows the BGCs detected in the genome of symbiotic and non-symbiotic Bacillus. b Reveals the variation of BGCs identified in symbiotic and non-symbiotic Bacillus. c Displays the difference between the numbers of antibiotic classes predicted from the BGCs of symbiotic and non-symbiotic Bacillus
Similar content being viewed by others
References
Kubanek J, Jensen PR, Keifer PA, Sullards MC, Collins DO, Fenical W (2003) Seaweed resistance to microbial attack: a targeted chemical defense against marine fungi. Proc Natl Acad Sci 100:6916–6921. https://doi.org/10.1073/pnas.1131855100
Blunt JW, Copp BR, Munro MHG, Northcote PT, Prinsep MR (2004) Marine natural products. Nat Prod Rep 21:1–49. https://doi.org/10.1039/b305250h
Satheesh S, Ba-akdah MA, Al-Sofyani AA (2016) Natural antifouling compound production by microbes associated with marine macroorganisms—a review. Electron J Biotechnol 21:26–35. https://doi.org/10.1016/j.ejbt.2016.02.002
Brinkmann CM, Marker A, Kurtböke Dİ (2017) An overview on marine sponge-symbiotic bacteria as unexhausted sources for natural product discovery. Diversity 9(4):40. https://doi.org/10.3390/d9040040
Wibowo JT, Kellermann MY, Versluis D, Putra MY, Murniasih T, Mohr KI, Wink J, Engelmann M, Praditya DF, Steinmann E, Schupp PJ (2019) Biotechnological potential of bacteria isolated from the sea cucumber Holothuria leucospilota and Stichopus vastus from Lampung. Indones Mar Drugs 17:635. https://doi.org/10.3390/md17110635
Hagstrom A, Pommier T, Rohwer F, Simu K, Stolte W, Svensson D, Zweifel UL (2002) Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl Environ Microbiol 68(7):3628–3633. https://doi.org/10.1128/AEM.68.7.3628-3633.2002
Debbab A, Aly AH, Lin WH, Proksch P (2010) Bioactive compounds from marine bacteria and fungi. Microb Biotechnol 3(5):544–563. https://doi.org/10.1111/j.1751-7915.2010.00179.x
Song Y, Cai ZH, Lao YM, Jin H, Ying KZ, Lin GH, Zhou J (2018) Antibiofilm activity substances derived from coral symbiotic bacterial extract inhibit biofouling by the model strain Pseudomonas aeruginosa PAO1. Microb Biotechnol 11(6):1090–1105. https://doi.org/10.1111/1751-7915.13312
Viju N, Ezhilraj N, Shankar CVS, Punitha SMJ, Satheesh S (2018) Antifouling activities of extracellular polymeric substances produced by marine bacteria associated with the gastropod (Babylonia sp.). Nova Biotechnol Chim 17(2):115–124. https://doi.org/10.2478/nbec-2018-0012
Naughton LM, Romano S, O’Gara F, Dobson ADW (2017) Identification of secondary metabolite gene clusters in the Pseudovibrio genus reveals encouraging biosynthetic potential toward the production of novel bioactive compounds. Front Microbiol 8:1494. https://doi.org/10.3389/fmicb.2017.01494
Miller I, Vanee N, Fong S, Lim-Fong G, Kwan J (2016) Lack of overt genome reduction in the bryostatin-producing Bryozoan symbiont “Candidatus Endobugula sertula”. Appl Environ Microbiol 82:6573–6583. https://doi.org/10.1128/AEM.01800-16
Schmidt EW, Nelson JT, Rasko DA, Sudek S, Eisen JA, Haygood MG, Ravel J (2005) Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci 102(20): 7315–7320 doi: https://doi.org/10.1073/pnas.0501424102
Mast Y, Weber T, Golz M, Ort-Winklbauer R, Gondran A, Wohlleben A, Schinko E (2011) Characterization of the ‘pristinamycin supercluster’ of Streptomyces pristinaespiralismbt. Microb Biotechnol 4(2):192–206. https://doi.org/10.1111/j.1751-7915.2010.00213.x
Viju N, Punitha SMJ, Satheesh S (2018) Antifouling properties of bacteria associated with marine oyster Crassostrea sp. Thalasses. https://doi.org/10.1007/s41208-018-0095-9
Chaston J, Goodrich-Blair H (2010) Common trends in mutualism revealed by model associations between invertebrates and bacteria. FEMS Microbiol Rev 34:41–58. https://doi.org/10.1111/j.1574-6976.2009.00193.x
Bernasconi R, Stat M, Koenders A, Paparini A, Bunce M, Huggett MJ (2019) Establishment of coral-bacteria symbioses reveal changes in the core bacterial community with host ontogeny. Front Microbiol 10:1529. https://doi.org/10.3389/fmicb.2019.01529
Egerton FN (2015) History of ecological sciences, part 52: symbiosis studies. Bull Ecol Soc Am 96(1):80–139
Singh RP, Reddy CRK (2014) Seaweed–microbial interactions: key functions of seaweed-associated bacteria. FEMS Microbiol Ecol 88:213–230. https://doi.org/10.1111/1574-6941.12297
Slattery M, Rajbhandari I, Wesson K (2001) Competition-mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microb Ecol 41:90–96. https://doi.org/10.1007/s002480000084
Lemos ML, Dopazo CP, Toranzo AE, Barja JL (1991) Competitive dominance of antibiotic-producing marine bacteria in mixed cultures. J Appl Bacterial 71:228–232. https://doi.org/10.1111/j.1365-2672.1991.tb04452.x
Dalisay DS, Williams DE, Wang XL, Centko R, Chen J, Andersen RJ (2013) Marine sediment-derived Streptomyces bacteria from British Columbia, Canbada are a promising microbiota resource for the discovery of antimicrobial natural products. PLoS ONE 8(10):e77078. https://doi.org/10.1371/journal.pone.0077078
Long RA, Azam F (2001) Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol 67(11):4975–4983. https://doi.org/10.1128/AEM.67.11.4975-4983.2001
Imamura N, Nishijima M, Takadera T, Adachi K (1997) New anticancer antibiotics pelagiomicins produced by a new marine bacterium Pleagiobacter variabilis. J Antibiot 50:8–12. https://doi.org/10.7164/antibiotics.50.8.s
Olguin-Uribe G, Abou-Mansour E, Boulander A, Debard H, Francisco C, Combaut G (1997) 6-Bromoindole-3-carbaldehyde, from an Acinetobacter sp. bacterium associated with the ascidian Stomoza murrayi. J Chem Ecol 23:2507–2521
Girao M, Ribeiro I, Ribeiro T, Azevedo IC, Pereira F, Urbatzka R, Leao PN, Carvalho MF (2019) Actinobacteria isolated from Laminaria ochroleuca: A source of new bioactive compounds. Front Microbiol 10:683. https://doi.org/10.3389/fmicb.2019.00683
Raimundo I, Silva SG, Costa R, Keller-Costa T (2018) Bioactive secondary metabolites from octocoral-associated microbes—new chances for blue growth. Mar Drugs 16(12):485. https://doi.org/10.3390/md16120485
Moghaddam AJ, Crusemann M, Alanjary M, Harms H, Davila-Cespedes A, Blom J, Poehlein A, Ziemert N, Konig GM, Schaberle TF (2018) Analysis of the genome and metabolome of marine Myxobacteria reveals high potential for biosynthesis of novel specialized metabolites. Sci Rep 8:16600. https://doi.org/10.1038/s41598-018-34954-y
Piel J, Butzke D, Fusetani N, Hui D, Platzer M, Wen G, Matsunaga S (2005) Exploring the chemistry of uncultivated bacterial symbionts: antitumor polyketides of the pederin family. J Nat Prod 68:472–481. https://doi.org/10.1021/np049612d
Agrawal S, Acharya D, Adholeya A, Barrow CJ, Deshmukh SK (2017) Nonribosomal peptides from marine microbes and their antimicrobial and anticancer potential. Front Pharmacol 8:828. https://doi.org/10.3389/fphar.2017.00828
Barsby T, Kelly MT, Gagné SM, Andersen RJ (2001) Bogorol A produced in culture by a marine Bacillus sp. reveals a novel template for cationic peptide antibiotics. Org Lett 3:437–440. https://doi.org/10.1021/ol006942q
Isnansetyo A, Cui L, Hiramatsu K, Kamei Y (2003) Antibacterial activity of 2,4-diacetylphloroglucinol produced by Pseudomonas sp. AMSN isolated from a marine alga, against vancomycin resistant Staphylococcus aureus. Int J Antimicrob Agents 22:545–547. https://doi.org/10.1016/s0924-8579(03)00155-9
Huang C, Yang C, Zhu Y, Zhang W, Yuan C, Zhang C (2018) Marine bacterial aromatic polyketides from host-dependent heterologous expression and fungal mode of cyclization. Front Chem 6:528. https://doi.org/10.3389/fchem.2018.00528
Bewley CA, Holland ND, Faulkner DJ (1996) Two classes of metabolites from Theonella swinhoei are localized in distinct populations of bacterial symbionts. Experientia 52:716–722. https://doi.org/10.1007/BF01925581
Feliatra F, Muchlisin Z, Teruna H, Utamy W, Nursyirwani N, Dahliaty A (2018) Potential of bacteriocins produced by probiotic bacteria isolated from tiger shrimp and prawns as antibacterial to Vibrio, Pseudomonas, and Aeromonas species on fish. F1000Res 7:415. https://doi.org/10.12688/f1000research.13958.1
Xue C, Tian L, Xu M, Deng Z, Lin W (2008) A new 24 membred lactone and new polyen lactone from the marine bacterium Bacillus marinus. J Antibiot 61:668–674. https://doi.org/10.1038/ja.2008.98
Gillor O, Etzion A, Riley MA (2008) The dual role of bacteriocins as anti- and probiotics. Appl Microbiol Biotechnol 81(4):591–606. https://doi.org/10.1007/s00253-008-1726-5
Sugita H, Matsuo N, Hirose Y, Iwato M, Deguchi Y (1997) Vibrio sp. strain NM 10, isolated from the intestine of a Japanese coastal fish, has an inhibitory effect against Pasteurella piscicida. Appl Environ Microbiol 63(12):4986–4989
Zai AS, Ahmad S, Rasool SA (2009) Bacteriocin production by indigenous marine catfish associated Vibrio spp. Pak J Pharm Sci 22:162–167. https://doi.org/10.1016/j.cbpa.2010.10.020
Amin SA, Green DH, Hart MC, Küpper FC, Sunda WG, Carrano CJ (2009) Photolysis of iron–siderophore chelates promotes bacterial–algal mutualism. Proc Natl Acad Sci 106(40):17071–17076. https://doi.org/10.1073/pnas.0905512106
Manwar AV, Khandelwal SR, Chaudhari JM, Meyer SB, Chincholkar SB (2004) Siderophore production by a marine Pseudomonas aeruginosa and its antagonistic action against phytopathogenic fungi. Appl Biochem Biotechnol 118:243–251. https://doi.org/10.1385/ABAB:118:1-3:243
Soengas RG, Larrosa M, Balado M, Rodriguez J, Lemos ML, Jimenez C (2008) Synthesis and biological activity of analogues of vanchrobactin, a siderophore from Vibrio anguillarum serotype O2. Org Biomol Chem 6:1278–1287. https://doi.org/10.1039/b719713f
Mansson M, Gram L, Larsen TO (2011) Production of bioactive secondary metabolites by marine Vibrionaceae. Mar Drugs 9:1440–1468. https://doi.org/10.3390/md9091440
Rath CM, Janto B, Earl J, Ahmed A, Hu FZ, Hiller L, Dahlgren M, Kreft R, Yu F, Wolff JJ, Kweon HK, Christiansen MA, Håkansson K, Williams RM, Ehrlich GD, Sherman DH (2011) Meta-omic characterization of the marine invertebrate microbial consortium that produces the chemotherapeutic natural product ET-743. ACS Chem Biol 6:1244–1256. https://doi.org/10.1021/cb200244t
Asano N (2003) Glycosidase inhibitors: update and perspectives on practical use. Glycobiology 13(10):93R-104R. https://doi.org/10.1093/glycob/cwg090
Waters AL, Peraud O, Kasanah N, Sims JW, Kothalawala N, Anderson MA, Abbas SH, Rao KV, Jupally VR, Kelly M, Dass A, Hill RT, Hamann MT (2014) An analysis of the sponge Acanthostronglophora igens microbiom yields an actinomycete that produces the natural product manzamine A. Front Mar Sci 1:54. https://doi.org/10.3389/fmars.2014.00054
Palomo S, González I, de la Cruz M, Martín J, Tormo JR, Anderson M, Hill RT, Vicente F, Reyes F, Genilloud O (2013) Sponge-derived Kocuria and Micrococcus spp. as new sources of thiazolyl peptide antibiotics. Mar Drugs 11(4):1071–1086. https://doi.org/10.3390/md11041071
Martin J, Sousa T, Crespo G, Palomo S, Gonzalez I, Tormo JR, De La Cruz M, Anderson M, Hill RT, Vicente F et al (2013) Kocurin the tru structure of PM 181104, an anti-MRSA thiazolyl peptide from the marine-derived bacterium Kocuria palustris. Mar Drugs 11:387–398. https://doi.org/10.3390/md11020387
Fischbach MA, Walsh CT (2006) Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev 106:3468–3496. https://doi.org/10.1021/cr0503097
Nakashima Y, Egami Y, Kimura M, Wakimoto T, Abe I (2016) Metagenomic analysis of the sponge discodermia reveals the production of the cyanobacterial natural product Kasumigamide by ‘Entotheonella’. PLOSONE 11:e0164468. https://doi.org/10.1371/journal.pone.0164468
Harjes J, Ryu T, Abdelmohsen UR, Moitinho-Silva L, Horn H, Ravasi T, Hentschel U (2014) Draft genome sequence of the antitrypanosomally active sponge-associated bacterium Actinokineospora sp. strain EG49. Genome Announc. https://doi.org/10.1128/genomeA.00160-14
Gavriilidou A, Mackenzie T, Sánchez P, Tormo J, Ingham C, Smidt H, Sipkema D (2021) Bioactivity screening and gene-trait matching across marine sponge-associated bacteria. Mar Drugs 19:1–17. https://doi.org/10.3390/md19020075
Stein T (2005) Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol 56:845–857. https://doi.org/10.1111/j.1365-2958.2005.04587.x
Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 25:447–453. https://doi.org/10.1038/nbt1297
Weber T, Rausch C, Lopez P, Hoof I, Gaykova V, Huson DH, Wohlleben W (2009) CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J Biotechnol 140:13–17. https://doi.org/10.1016/j.jbiotec.2009.01.007
Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R (2011) AntiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:W339–W346. https://doi.org/10.1093/nar/gkr466
Tran PN, Yen MR, Chiang CY, Lin HC, Chen PY (2019) Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi. Appl Microbiol Biotechnol 103:3277–3287. https://doi.org/10.1007/s00253-019-09708-z
Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SJ, Suarez Duran HG, Los Santos ELC, Kim HU, Nave M et al (2017) AntiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45:W36–W41. https://doi.org/10.1093/nar/gkx319
Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Muller R, Wohlleben W, Breitling R, Takano E, Medema MH (2015) AntiSMASH 30-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43(W1):W237–W243. https://doi.org/10.1093/nar/gkv437
Van Heel AJ, de Jong A, Montalban-Lopez M, Kok J, Kuipers OP (2013) BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res 41:W448–W453. https://doi.org/10.1093/nar/gkt391
Prihoda D, Maritz JM, Klempir O, Dzamba D, Woelk CH, Hazuda DJ, Bitton DA, Hannigan GD (2021) The application potential of machine learning and genomics for understanding natural product diversity, chemistry, and therapeutic translatability. Nat Prod Rep. https://doi.org/10.1039/D0NP00055H
Belknap KC, Park CJ, Barth BM, Andam CP (2020) Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci Rep 10:2003. https://doi.org/10.1038/s41598-020-58904-9
Uria A (2012) Investigating natural product biosynthesis in uncultivated symbiotic bacteria of the marine sponge Theonella swinhoei. Hokkaido University, Sapporo
Dahiya R, Dahiya S, Fuloria NK, Kumar S, Mourya R, Chennupati SV, Jankie S, Gautam H, Singh S, Karan SK, Maharaj S, Fuloria S, Shrivastava J, Agarwal A, Singh A, Kishor A, Jadon G, Sharma A (2020) Natural bioactive thiazole-Based peptides from marine resources: structural and pharmacological aspects. Mar Drugs 18:329. https://doi.org/10.3390/md18060329
Beleneva IA (2008) Distribution and characterisics of Bacillus bacteria associated with hydrobionts and the waters of the Peter the Great Bay, Sea of Japan. Microbiology 77:497–503. https://doi.org/10.1134/S0026261708040176
Viju N, Punitha SMJ, Satheesh S (2019) Antifouling potential of symbiotic marine bacterium Bacillus subtilis MUT:M15 associated with the cuttlefish Sepia sp. Indian J Geo-Mar Sci 48(05):685–697
Viju N, Satheesh S, Punitha SMJ (2020) Antibiofilm activity of symbiotic Bacillus species associated with marine gastropods. Ann Microbiol. https://doi.org/10.1186/s13213-020-01554-z
McCutcheon JP, Moran NA (2011) Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. https://doi.org/10.1038/nrmicro2670
Waters DJ (2018) Toxins with potential for drug development. Mar Drugs 16:162. https://doi.org/10.3390/md16050162
Hirose E, Neilan BA, Schmidt EW, Murakami A (2009) Enigmatic life and evolution of Prochloron and related cyanobacteria inhabiting colonial ascidians. In: Gault PM, Marler HJ (eds) Handbook on cyanobacteria: biochemistry, biotechnology and applications. Nova Publishers, New York, pp 161–189
Nakabachi A, Ueoka R, Oshima K, Teta R, Mangoni A, Gurgui M, Oldham NJ, van Echten-Deckert G, Okamura K, Yamamoto K, Inoue H, Ohkuma M, Hongoh Y, Miyagishima SY, Hattori M, Piel J, Fukatsu T (2013) Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol 23:1478–1484. https://doi.org/10.1016/j.cub.2013.06.027
Schmidt EW (2015) The secret to a successful relationship: lasting chemistry between ascidians and their symbiotic bacteria. Invertebr Biol 134(1):88–102. https://doi.org/10.1111/ivb.12071
Kwan JC, Donia MS, Han AW, Hirose E, Haygood MG, Schmidt EW (2012) Genome streamlining and chemical defense in a coral reef symbiosis. Proc Natl Acad Sci 109(50):20655–20660. https://doi.org/10.1073/pnas.1213820109
Webster N, Hill R (2001) The culturable microbial community of the great barrier reef sponge Rhopaloeides odorabile is dominated by an α-Proteobacterium. Mar Biol 138:843–851. https://doi.org/10.1007/s002270000503
Linneman J, Paulus D, Lim-Fong G, Lopanik NB (2014) Latitudinal variation of a defensive symbiosis in the Bugula neritina (Bryozoa) sibling species complex. PLOSONE 9(10):e108783. https://doi.org/10.1371/journal.pone.0108783
Enticknap JJ, Kelly M, Peraud O, Hill RT (2006) Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl Environ Microbiol 72:3724–3732. https://doi.org/10.1128/AEM.72.5.3724-3732.2006
Crowley S, O’Gara F, O’Sullivan O, Cotter P, Dobson A (2014) Marine Pseudovibrio sp. as a novel source of antimicrobials. Mar drugs 12:5916–5929. https://doi.org/10.3390/md12125916
Harrington C, Reen FJ, Mooij MJ, Stewart FA, Chabot JB, Guerra AF, Glöckner FO, Nielsen KF, Gram L, Dobson ADW, Adams C, O’Gara F (2014) Characterisation of non-autoinducing tropodithietic acid (TDA) production from marine sponge Pseudovibrio species. Mar Drugs 12(12):5960–5978. https://doi.org/10.3390/md12125960
Penesyan A, Tebben J, Lee M, Thomas T, Kjelleberg S, Harder T, Egan S (2011) Identification of the antibacterial compound produced by the marine epiphytic bacterium Pseudovibrio sp. D323 and related sponge-associated bacteria. Mar Drugs 9:1391–1402. https://doi.org/10.3390/md9081391
Bondarev V, Richter M, Romano S, Piel J, Schwedt A, Schulz-Vogt HN (2013) The genus Pseudovibrio contains metabolically versatile bacteria adapted for symbiosis. Environ Microbiol 15(7):2095–2113. https://doi.org/10.1111/1462-2920.12123
Versluis D, Nijsse B, Naim MA, Koehorst JJ, Wiese J, Imhoff JF, Schaap PJ, van Passel MWJ, Smidt H, Sipkema D (2018) Comparative genomics highlights symbiotic capacities and high metabolic flexibility of the marine genus Pseudovibrio. Genome Biol Evol 10(1):125–142. https://doi.org/10.1093/gbe/evx271
Romano S, Jackson SA, Patry S, Dobson ADW (2018) Extending the “One Strain Many Compounds” (OSMAC) principle to marine microorganisms. Mar Drugs 16(7):244. https://doi.org/10.3390/md16070244
Adnani N, Rajski SR, Bugni TS (2017) Symbiosis-inspired approaches to antibiotic discovery. Nat Prod Rep 34(7):784–814. https://doi.org/10.1039/c7np00009j
Rosenberg IZ, Rosenberg E (2008) Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32(5):723–735. https://doi.org/10.1111/j.1574-6976.2008.00123.x
Bachmann BO, Van Lanen SG, Baltz RH (2014) Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J Ind Microbiol Biotechnol 41(2):175–184. https://doi.org/10.1007/s10295-013-1389-9
Baral B, Akhgari A, Metsa-Ketela M (2018) Activation of microbial secondary metabolic pathways: avenues and challenges. Synth Syst Biotechnol 3(3):163–178. https://doi.org/10.1016/j.synbio.2018.09.001
Alvarez-Alvarez R, Martínez-Burgo Y, Rodríguez-García A, Liras P (2017) Discovering the potential of S.clavuligerus for bioactive compound production: cross-talk between the chromosome and the pSCL4 megaplasmid. BMC Genom 18:907. https://doi.org/10.1186/s12864-017-4289-y
Reen FJ, Romano S, Dobson ADW, O’Gara F (2015) The sound of silence: activating silent biosynthetic gene clusters in marine microorganisms. Mar Drugs 13(8):4754–4783. https://doi.org/10.3390/md13084754
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A et al (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. https://doi.org/10.1038/417141a
Tambadou F, Lanneluc I, Sable S, Klein GEL, Doghri I, Sopena V, Didelot S, Barthelemy C, Thiery VE, Chevrot R (2014) Novel nonribosomal peptide synthetase (NRPS) genes sequenced from intertidal mudflat bacteria. FEMS Microbiol Lett 357:123–130
Katz L, Baltz RH (2016) Natural product discovery: past, present, and future. J Ind Microbiol Biotechnol 43:1551–1576. https://doi.org/10.1007/s10295-015-1723-5
Winter JM, Behnken S, Hertweck C (2011) Genomics-inspired discovery of natural products. Curr Opin Chem Biol 15:22–31. https://doi.org/10.1016/j.cbpa.2010.10.020
Rutledge PJ, Challis GL (2015) Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol 13:509–523. https://doi.org/10.1038/nrmicro3496
Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C (2007) Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol 3:213–217. https://doi.org/10.1038/nchembio869
Bode HB, Bethe B, Hofs R, Zeeck A (2002) Big effects from small changes: possible ways to explore nature’s chemical diversity. Chem BioChem 3:619–627. https://doi.org/10.1002/1439-7633(20020703)3:7%3c619::AID-CBIC619%3e3.0.CO;2-9
Nutzmann HW, Reyes-Dominguez Y, Scherlach K, Schroeckh V, Horn F, Gacek A et al (2011) Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proc Natl Acad Sci 108:14282–14287. https://doi.org/10.1073/pnas.1103523108
Netzker T, Fischer J, Weber J, Mattern DJ, König CC, Valiante V, Brakhage AA (2015) Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front Microbiol 6:299. https://doi.org/10.3389/fmicb.2015.00299
Rateb ME, Houssen WE, Harrison WTA, Deng H, Okoro CK, Asenjo JA, Andrews BA, Bull AT, Goodfellow M, Ebel R et al (2011) Diverse metabolic profiles of a Streptomyces strain isolated from a hyper-arid environment. J Nat Prod 74:1965–1971. https://doi.org/10.1021/np200470u
Metsa-Ketela M, Ylihonko K, Mäntsala P (2004) Partial activation of a silent angucyclinetype gene cluster from a rubromycin beta producing Streptomyces sp. PGA64. J Antibiot 57:502–510. https://doi.org/10.7164/antibiotics.57.502
Egli T (2015) Microbial growth and physiology: a call for better craftsmanship. Front Microbiol 6:287. https://doi.org/10.3389/fmicb.2015.00287
Funding
This research did not receive any specific Grant from funding agencies in the public, commercial, or nonprofit sectors.
Author information
Authors and Affiliations
Contributions
All the authors of this manuscript contributed equally. Also, all the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest has been declared by the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Viju, N., Punitha, S.M.J. & Satheesh, S. An Analysis of Biosynthesis Gene Clusters and Bioactivity of Marine Bacterial Symbionts. Curr Microbiol 78, 2522–2533 (2021). https://doi.org/10.1007/s00284-021-02535-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02535-4