Abstract
Aquaponic systems are sustainable solutions for food production combining fish growth (aquaculture) and production of vegetables (hydroponic) in one recirculating system. In aquaponics, nitrogen-enriched wastewater from fish in the aquaculture serves as fertilizer for the plants in the hydroponics, while the nitrogen-depleted and detoxified water flows back to the aquaculture. To investigate bacterial nitrogen-cycling in such an aquaponic system, measurements of nitrogen species were coupled with time-resolved 16S rRNA gene profiling and the functional capacity of organisms was studied using metagenomics. The aquaponic system was consistently removing ammonia and nitrite below 23 µM and 19 µM, and nitrate to steady-state concentrations of about 0.5 mM. 16S rRNA gene amplicon sequencing of sediments exposed in the pump sump revealed that typical signatures of canonical ammonia-oxidising microorganisms were below detection limit. However, one of the most abundant operational taxonomic units (OTU) was classified as a member of the genus Nitrospira with a relative abundance of 3.8%. For this genus, also genome scaffolds were recovered encoding the only ammonia monooxygenase genes identified in the metagenome. This study indicates that even in highly efficient aquaponic systems, comammox Nitrospira were found to participate in ammonium removal at low steady-state ammonia concentrations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the last 50 years, aquaponic systems have become a promising biotechnology for sustainable food production. In aquaponics, aquaculture (fish cultivation) and hydroponics (vegetables production) are integrated into one water-circulating system. One of the main issues in aquaculture is the accumulation of ammonium due to decomposed fish food and fish excrements. Depending on the fish species and the exposure time [1], ammonia is chronically toxic to fish in concentrations higher than 1.9 mg/L total ammonia nitrogen at pH 7 and 20 °C. For the common carp (Cyprinus carpio), it was shown that even an ammonia concentration of 1.3 mg/L can be mortal to small individuals of about 10 g [2]. Therefore, the water of aquaculture systems must be replaced regularly or requires cleaning. Aquaponic systems solve the issue of regeneration by pumping water from the fish tank into the grow beds of the hydroponic part, where it serves as fertilizer for vegetables, fruits, or herbs. Plants can use both ammonium and nitrate as nitrogen sources [3]. However, for lettuce, pak choi, tomato, and chive, it was shown that nitrogen is mainly assimilated in the form of nitrate [4].
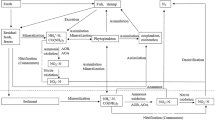
The essential reaction in the aquaponic system is the nitrification during which toxic ammonium is oxidised to less harmful nitrate, which can be carried out by different microorganisms [5, 6]. It was shown that low pH can shift microbial communities in an aquaponic system [7]. Since the complete oxidation of ammonia to nitrate is an essential step for the effective operation in an aquaponic system, the nitrogen turnover in such a system was elucidated in this study.
Until 2015, nitrification was assumed to be exclusively performed in a two-step process carried out by two phylogenetically distinct bacterial lineages. In the first step, ammonium is oxidised to nitrite via hydroxylamine by e.g. Nitrosomonas. In the second step, nitrite is oxidised to nitrate by e.g. Nitrobacter or Nitrospira. This process was observed in marine [8, 9] and freshwater [10,11,12,13] aquaponic or aquaculture systems by enrichment techniques, fluorescence in situ hybridization, or sequence analyses.
In 2015, Daims et al. [14] and van Kessel et al. [15] discovered that nitrification can also be carried out by one single organism affiliated to the genus Nitrospira. By showing that this bacterium was capable of oxidising ammonium fully to nitrate, a process called ‘comammox’ (complete ammonia oxidation), the authors overturned the 100-yr-old lasting paradigm that nitrification can only be performed by two distinct groups of organisms. Since their discovery, comammox Nitrospira have been frequently detected in aquifers [16], drinking water systems [17], wastewater treatment plants [18], as well as in recirculating aquaculture systems [19]. Furthermore, 16S rRNA gene analysis of community compositions in different compartments of an aquaponic system showed that nitrification took place on a biofilter located behind the fish tank retaining large particle matter [20]. Since Nitrospira was among the most abundant species and other nitrifying bacteria seemed not to be present in the biofilm community of the aquaponic system, the authors assumed that the nitrification process was carried out by Nitrospira alone.
So far, all known comammox Nitrospira belong to the sublineage II of Nitrospira [14, 15, 17], which comprises comammox Nitrospira species and nitrite-oxidising (canonical) Nitrospira species [21, 22]. Additionally, comammox and canonical Nitrospira form mixed phylogenetic clades within this sublineage [14, 15, 17], suggesting that they cannot be distinguished based on 16S rRNA sequences alone.
Here, we aim at elucidating the type of microbial nitrogen metabolism in an aquaponic system and how the microbial communities develop over time. A combination of 16S rRNA gene sequencing and metagenomic analysis is employed for distinguishing different types of nitrification and to follow the respective functional clades.
Material and Methods
The Aquaponic System
The aquaponic system investigated in this study was a private backyard system. The fish tank was built from a regular 1200 L plastic intermediate bulk container (IBC) and filled with 1000 L tap water (Fig. 1). The hydroponic part consisted of two grow beds, each 100 × 120 cm in size and filled with gravel up to 30 cm in height. The overflow water from the fish tank flew into the grow beds by gravity and was treated by a biofilter made of gauze with 1 mm mesh size to remove larger particles. The particle filter was cleaned daily. The water of the grow beds was periodically released into a sump by a hydraulic siphon system. From there, it was continuously pumped back into the fish tank with an adjusted flow rate of 800 L/h. The aquaponic system was installed in the open and environmental conditions such as exposure to sunlight and temperature were not controlled. Water losses due to evapotranspiration were compensated by refilling with tap water.
In the beginning of the operation, five common carps (Cyprinus carpio) and five wels catfish (Silurus glanis) were grown corresponding to a fish density of 10 fish/cm3. After a fish disease killing all the fish, the water in the aquaponic system was completely exchanged and all system parts were cleaned. Only ten carps were grown subsequently resulting in individuals of 400 g weight each at the end of the experiment. The fish were fed with dried honey bee larvae. Four Lettuce, three tomatoes, two cucumber, four chard, and three strawberries were grown in the grow beds from May to October.
Preparation of Test Sediments and Sampling
Quartz sand (1 mm grain size) was autoclaved for 20 min at 120 °C and introduced into the pump sump as substratum for microbial growth. Regular plastic hair curlers (6.5 cm in length) with porous walls (1 × 0.5 mm pore size) were filled with approximately 10 g of the sand and closed with parafilm at both ends. The test sediments were incubated in the aquaponic sump. In total, 12 curlers were taken for amplicon sequencing over a period of 508 days ending in October. For metagenome sequencing, DNA was extracted from one test sediment taken at the last day of incubation. Water was sampled on all sampling days to check the system performance and analyses of nitrogen species concentrations (see below). The pH was determined onsite using test stripes (pH 4.5–10, Carl Roth GmbH, Karlsruhe, Germany) to guarantee a steady pH over time.
Chemical Analysis
Water samples were regularly taken from the aquaponic sump and concentrations of nitrate, nitrite, and ammonium were analysed in triplicates with ion chromatography. The samples were prepared by 1:2 dilution with 0.01 mM potassium buffer for the analysis of anions and with 20 mM methyl sulfonic acid (70% v/v) (Merck KGaA, Darmstadt, Germany) for the analysis of cations. All samples were centrifuged for 15 min at 16,000×g to remove solids. 200 µL of the supernatants were diluted 1:5 with ultrapure water (18.2 µS, Merck Millipore System, Merck KGaA, Darmstadt, Germany). The samples were stored at − 20 °C until analysis.
Anion and cation concentrations were measured with an ion chromatograph (Thermo Fisher Scientific, Dreieich, Germany) equipped with a Dionex™ IonPac™ AS23-4 µm column and an AERS 500 suppressor (2 mm) for the measurement of anions using 0.8 mM NaHCO3/4.5 mM Na2CO3 (Thermo Fisher Scientific, Dreieich, Germany) as eluent at a flow rate of 0.25 mL/min at 7 mA. Ammonium was measured with a Dionex™ IonPac™ CS12 A column and a CERS 500 suppressor (2 mm) using 20 mM methyl sulfonic acid (70% v/v) (Merck KGaA, Darmstadt, Germany) as eluent at a flow rate of 0.25 mL/min at 15 mA. The detection limit was calculated based on the calibration method according to DIN EN 32645. Non-equidistant calibration points (10, 20, 50, 100, 200 µM) were used.
DNA Extraction
DNA was extracted from the sand of each curler using the FastDNA™ SPIN Kit for Soil (MP Biomedicals, Heidelberg, Germany). 460–480 mg of sand (wet weight) were taken and treated as described in the manufacturers’ instructions. For the cell lysis, the bead-beating system Precellys24 tissue homogenizer [Bertin Instruments, Montigny-le-Bretonneux, France] was used for two times at 65,000 rpm for 30 s each turn. The DNA samples were stored at − 20 °C until further usage.
Preparation of 16S rRNA Gene Amplicon Library
The preparation of the amplicon library was adapted from the Illumina 16S sequencing library preparation guide (part no. 15044223 Rev. B). The primers Pro341F/Pro805R [23] targeting the V3-V4 region of 16S rRNA genes of bacteria and archaea were applied to get 250 bp reads lengths. They were combined with the Illumina overhang adapters (Eurofins Genomics, Ebersberg, Germany).
For the first stage PCR, 2 µL of extracted DNA was mixed with 1X KAPA HiFi Hot Start Ready Mix (Roche, Basel, Switzerland), 0.25 μM of each the forward and the reverse primers including the Illumina overhang adapters, and nuclease-free water (Qiagen, Hilden, Germany) to a final reaction volume of 25 µL. Duplicates were prepared for each sample and pooled after the PCR. The PCR amplification was carried out with an initial denaturation step at 94 °C for 5 min followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 70 °C for 1 min, and a final extension at 70 °C for 5 min.
The PCR amplicons were purified using MagSi-NGSPREP Plus magnetic beads (Steinbrenner, Wiesenbach, Germany) by thoroughly mixing 32 µL of magnetic beads with 40 µL of samples and following the PCR clean-up instructions given in the Illumina 16S metagenomic sequencing library preparation guide with the exception that the beads were resuspended in 42.5 µL of elution buffer EB (Qiagen, Hilden, Germany). 40 µL of the supernatants were then taken for further analyses.
The index PCR was performed using the Nextera XT DNA Library Preparation Kit v2 Set D (FC-131-2004) from Illumina (Munich, Germany). The PCR and the second PCR clean-up were performed as described in the Illumina 16S metagenomic sequencing library preparation guide.
DNA concentrations were measured with a Qubit fluorometer using the Qubit™ dsDNA HS Assay Kit (ThermoFisher Scientific, Dreieich, Germany). The samples were normalised to a concentration of 2 ng/µL and 5 µL of all samples were combined into one ready-to-load sample containing 12 libraries, which was analysed by GATC Biotech AG (Konstanz, Germany) on an Illumina Miseq platform. The 16S rRNA gene sequence reads are deposited in the NCBI’s nr database as SUB5504898 in the bioproject PRJNA534201.
Analysing 16S rRNA Gene Sequencing Data
The 16S rRNA sequences were analysed as paired-end run using the mothur-based MetaAmp Version 2.0 software [24]. The settings were similarity cutoff of 0.97, minimum overlap of 35 bp, no mismatches in the overlap region, no differences in primer sequences, max. one expected error, and a trim amplicon length of 350 bp. The alignment of reads was conducted using the SILVA 128 database.
Statistical analyses were carried out using the R code described in [25]. For multivariate statistics, data were rarefied (to the lowest number of sequences in the samples, which was 173,469 reads per sample) and a Bray–Curtis distance was calculated. In order to account for rarefication biases, this procedure was repeated 100 times and the distance across these iterations was averaged. Alpha diversity was determined using the Shannon index. Principle coordinate analysis (PCoA) was used to display the beta diversity of the samples. Relationships between OTUs and environmental factors (time, pH, nitrate, ammonia, sulphate, chloride, sodium, magnesium, and calcium) were calculated using PERMANOVA (Adonis testing).
Identification of OTUs that significantly correlated with time in relative abundance were selected by applying a Pearson correlation. Only OTUs with a P-value < 0.001 are reported in the manuscript.
Metagenomic Sequencing and Analysis
To study functionalities, 390 ng of DNA were extracted from the sand sample on the last sampling day for the whole metagenome sequencing. Library preparation and Illumina HiSeq sequencing of paired-end 150-bps reads were done by GATC Biotech AG (Konstanz, Germany). Raw reads obtained from GATC Biotech were trimmed and quality filtered using bbduk (http://jgi.doe.gov/data-and-tools/bbtools/) and SICKLE version 1.21 (https://github.com/najoshi/sickle). The processed reads were assembled and scaffolded using metaSPADES version 3.10.1 [26]. For scaffolds longer than 1 kb genes were predicted using prodigal [27] and diamond blastp [28] was used to annotate the genes against the Unifref100 database [29], which contained taxonomy information from UniProt and the NCBI taxonomy database.
Databases of 100 amino acid sequences each for ammonia monooxygenase subunit A (amoA), subunit B (amoB), and subunit C (amoC) and the hydroxylamine reductase were created from highly identical sequences derived from NCBI’s nr database. The sequences were aligned using MUSCLE [30] and maximum likelihood phylogenetic trees were constructed based on the JTT matrix-based model [31] using the MEGA7 software [32] by applying default settings.
Results
Nitrogen Species in the Aquaponic System
The aquaponic system efficiently removed nitrogen from the water. Only nitrate was measurable in low concentrations up to 1.1 mM in the sump of the aquaponic system at the end of the system operation (Fig. 2). Ammonium and nitrite concentrations were always below 23 µM and 19 µM, respectively, indicating that both ammonium and nitrate were taken up by the plants and residual ammonium was completely oxidised by nitrifying microorganisms. In April 2017, the concentration of nitrate raised to 0.6 mM due to the starting metabolism of the fish resulting in a higher nitrogen load of the water. When seedlings were planted in May, nitrate and potentially ammonium were increasingly taken up by the plants leading to a decrease of nitrate concentrations to 0.1 mM in June 2017. The nitrate concentrations increased again from June onwards to 1.1 mM at the end of the operation in October 2017. The absence of ammonium in all samples indicated that a nitrifying microbial community was established in the aquaponic system. The pH stayed constant between 6.8 and 7 and was not adjusted.
Microbial Diversity
Sequencing of 16S rRNA genes revealed a highly diverse community structure in the aquaponic system. A total of 40 bacterial phyla were identified, out of which 15 made up 95.4% of the total microbial community (Supplementary material Figure S1). The most dominant phyla in the aquaponic system were Proteobacteria, Bacteroidetes, Verrucomicrobia, Acidobacteria, Actinobacteria, and Nitrospira. The three most abundant OTUs classified on family level at the end of the operation of the aquaponic system (day 508) were Verrucomicrobiaceae (4.3%), Nitrospiraceae (3.8%), and Comamonadaceae (2.8%), which belong to the phylum Proteobacteria.
In the highly diverse samples, the 100 most abundant genera make up almost 50% of the total community at the end of the operation of the aquaponic system (Fig. 3). After a water change on day 293, unclassified Verrucomicrobia, unclassified Chloroplasts, as well as Nitrospira, Aquabacterium, and Arenimonas were among the most dominant genera based on OTUs. The dominant OTU classified as Nitrospira was found in all samples after the water change. The classical genus of ammonia oxidisers, Nitrosomonas, was only found in very low relative abundance (0.03%) suggesting that the detected Nitrospira were fully oxidising ammonium to nitrate similar to the previously found Candidatus Nitrospira inopinata [14, 15].
Relative abundances of the 100 most abundant taxonomic genera in the aquaponic system based on 16S rRNA gene sequencing. Taxonomy was sorted based on the total reads of the sample “day 508”. Please consider the repetition of colours. A water exchange was performed on day 293. Other bacteria summarise all bacteria that could be classified but do not belong to the 15 most abundant bacterial taxa or Archaea. Unknown depicts all putative OTUs that could not be assigned to known taxa
The microbial diversity in the aquaponic system was described using the Shannon index HS (Fig. 4a). During the first year of operation, the diversity was increasing over time from 4.74 (day 9) to 6.30 (day 307). During the second year, the Shannon index stagnated between 6.27 and 6.39 indicating a stabilisation of the microbial diversity on a very high diversity level. A total water exchange was performed on day 293.
Shannon indices (a) as measure for microbial alpha diversity calculated from OTU abundances at different time points. Principal coordinate analysis diagram (b) showing the beta diversity of OTUs in the microbial community at different sampling days. Principal coordinate analysis axis 1 and axis 2 explained 44.2% and 22.5% of the total variance in the community, respectively
Principal coordinate analysis of the relative abundances of OTUs showed a distinct shift in the microbial community composition over time (Fig. 4b), which can be clustered in early, mid, and late time samples. Samples taken in the beginning of the operation in spring 2016 (day 9 and day 13) differed significantly from the samples taken in the midpoint of the operation at the end of summer (day 102 and day 115). After the change of water in spring 2017 (day 293), the late time cluster microbial community composition deviated again showing a precedent shift in the winter period. In the last half of the year of operation, the community composition seemed to become more stable and shifted only slightly with seasons.
As indicated in the principal coordinate analysis, time had a substantial impact on the microbial community composition (PERMANOVA P-value 0.001). Interestingly, no other measured parameter showed a significant association with the observed microbiome structure. OTUs classified as members of the genera Nitrospira, Acidobacteria, Sphingobacteria, and Cytophagia were less abundant or not even detectable in the beginning and increased continuously during the operation time of the aquaponic system (Fig. 3). Other genera, mainly belonging to the Alphaproteobacteria, decreased in abundances over time. However, even after one and a half years of operation, a steady-state community had not been reached according to the PERMANOVA test.
Metagenome Analyses
Metagenome sequencing was performed from the samples taken at the last sampling day (day 508) to investigate the metabolism of the key players in ammonia oxidation. The assembled metagenome showed that the ammonia monooxygenases genes amoA, amoB, and amoC were only found on scaffolds classified as Nitrospira. The ammonia monooxygenase subunits amoA (Fig. 5), amoB, and amoC (Supplementary material Fig. S3 and S4) showed highly identical amino acid sequences to those obtained from reference sequences of comammox Nitrospira nitrificans (Table 1). The sequences of amino acids showed highest identities (98% for amo subunits and 96% for hydroxylamine reductase) with comammox Nitrospira species found on a rapid sand filter and a household sand filter [33, 34]. Two further scaffolds contained amoA and amoC genes, respectively (Supplementary material Table S1).
Phylogenetic tree of amoA gene sequences using the maximum likelihood method based on the JTT matrix-based model [31]. Amo ammonia monooxygenase. Mmo methane monooxygenase. Pmo particulate methane monooxygenase. Scale bar indicates estimated number of substitutions per site
Moreover, hydroxylamine reductases were only found in Nitrospira (Supplementary material Figure S5). Two additional genes were identified for a putative hydroxylamine reductase (Supplementary material Table S1). Consequently, Nitrospira was the only significantly abundant genus capable of ammonia oxidation in the backyard aquaponic system.
Discussion
Performance of the Aquaponic System
The free ammonia concentration is one of the most important parameters in aquaculture since it is toxic to fish species in relatively low concentrations (1.3 mg/L for common carps) [2]. In an aquaponic system, free ammonia nitrogen is released into the tank by the fish and taken up by the plants in the grow beds either as ammonium or after oxidation to nitrate [3]. The nitrifying microbial community in the sediments converts the residual ammonium that is not taken up by plants to nitrate, which is less harmful to fish species than ammonia [5, 6]. Although nitrification and degradation of organic carbon lead to acidification, the gravel of the grow beds obviously buffered the system well enough to keep the pH constant. The crucial concentration of ammonium is at the end of the hydroponic part at the influent to the aquaculture fish tank. In our aquaponic system, a nitrifying community was efficiently established reducing the ammonium concentration effectively below 23 µM. However, the nitrate concentration is rising during summer and the begin of autumn exceeding the concentration of 23 µM. It seems that more ammonium was produced due to the fish growth and less nitrogen was assimilated into the plants, which were at their end phase of growth and food production. The surplus of ammonium was then converted most likely by comammox Nitrospira. These results can be underlined by the fact that the abundances of OTUs assigned to the genera Nitrospira correlated positively with time meaning they became more abundant during the end of the operation of the aquaponic system.
Nitrification in aquaculture biofilters is most efficient at slightly alkaline pH of 7.5 to 9.0 [35, 36]. However, hydroponic plants grow best at slightly acidic conditions such as pH 5.5 for romaine lettuce crops [37] and 5.5 to 6.0 for greenhouse cucumber [38]. Compromising the efficiencies of nitrification, fish cultivation and plant growth, neutral pH 7 is commonly accepted as the optimal pH for aquaponic systems [39]. Our open aquaponic system was continuously running at pH 6.8–7.0. The steady pH may arise from the high adjusted flow rate of 800 L/h so that ammonium could not accumulate in the fish tank. An adjustment was not needed providing an efficiently self-regulated system with optimal nitrogen removal.
Microbial Community Composition
Since the focus was on microbial nitrification processes, the test sediments installed in the pump sump were located between the grow beds and the fish tank. Until now, microbial diversity in aquaculture and aquaponic systems was mainly studied on biofilters [11, 12, 19, 20, 40]. In aquaponic systems, however, the biofilter function is replaced by the grow beds.
In our test sediments, the most abundant OTUs were member of Proteobacteria, Bacteroidetes, Verrucomicrobia, Acidobacteria and Actinobacteria. The same phyla were also among the most dominant ones of an aquaponics biofilter community [20]. In contrast, Bartelme et al. [19] identified Actinobacteria, Gammaproteobacteria, Planctomycetes, and Sphingobacteria as the most dominant phyla in freshwater recirculating aquaculture systems (RAS). Sugita et al. [12] found Alphaproteobacteria, Betaproteobacteria, Nitrospira, Actinobacteria, Bacilli, Gammaproteobacteria, Planctomycetacia, and Sphingobacteria as predominant phyla in similar systems. However, nitrification also takes place in the grow beds, which was not assessed here.
Surprisingly, the microbial community compositions in the aquaponic system changed continuously over 508 days. Since the parameters determined for the aquaponic system were relatively constant, the change of microbial community compositions only correlated with the factor time. However, this study focusses on the nitrification in an aquaponic system. The role of other bacteria that correlate with the factor time remains unclear.
There was no correlation detectable between the nitrifying microorganisms and nitrate concentrations. Since the ammonium concentration was always very small, the removal of the nitrogen compounds by the plants was in equilibrium with the ammonia production by the fish and the microbial nitrification keeping all nitrogen species at low steady-state concentrations.
Two-Step Nitrification or Comammox?
The microbial community composition on the sediments revealed Nitrospira as the only organisms potentially involved in nitrification. The classically known members of the genus Nitrospira perform the second part of the two-step nitrification, the oxidation of nitrite to nitrate. For a complete nitrification process, a second organism such as Nitrosomonas would be essential to oxidise ammonia to nitrite. However, ammonia-oxidising bacteria like Nitrosomonas were at negligible abundance in our aquaponic sediments. Nitrospira not only belonged to the most abundant organisms in the aquaponic system but was also most likely performing the complete ammonia oxidation to nitrate. Similar results were reported by Schmautz et al. [20], who found Nitrospira to be one of the most abundant species in the aquaponic system. Since other ammonia-oxidising bacteria were only found at very low abundance, the authors assumed that the detected Nitrospira were comammox organisms [20].
Ammonia is first oxidised to hydroxylamine by ammonia monooxygenase, which consists of at least three subunits (amoA, amoB, and amoC). Hydroxylamine is then oxidised to nitrite catalysed by the hydroxylamine reductase. These genes can, therefore, be taken as indicators for ammonia oxidation. Detection of both genes for ammonia and nitrite oxidation in one genome are strongly indicative of comammox [14]. In our aquaponic system, the scaffolds of genes for the ammonia-oxidising enzymes were only found on scaffolds classified as Nitrospira. Nitrosomonas und Nitrobacter, which have been regarded as the main nitrifying bacteria so far, do probably not play a pivotal role in nitrification in our freshwater aquaponic systems. Hence, the interpretation of the community analysis was strongly supported by the metagenomic analysis and the detection of genes coding for enzymes involved in nitrification.
Comammox Nitrospira were predicted to survive in environments with low ammonium concentrations [41]. This assumption was recently underlined, when comammox Nitrospira were detected in groundwater [16], drinking water systems [17, 33] as well as recirculating aquaculture systems with low ammonium loading [19]. Nitrification allows for only little energy conservation requiring a high substrate turnover for microbial growth [41]. Hence, the cells have to produce lots of enzymes dedicated to nitrification and fit those into the limited cell volume. At high ammonia concentrations and high growth rates, it might become beneficial that the whole nitrification process is shared as partial processes between the ammonium oxidizer and the nitrate oxidizer because they can produce higher rates if they fill the limited cell volume with the enzymes for only one partial process. The overall rate is then higher. At very low ammonia concentrations, however, comammox bacteria obtain an advantage because they grow at low rates, do not need that many enzymes, and can thus fit both processes for complete oxidation into one cell [41].
Due to the high similarities of the genes involved in complete ammonia oxidation between known comammox Nitrospira and those obtained in this study, their high abundance, and the non-detection of classical ammonia oxidizers, we conclude that ammonia was mainly oxidised by comammox Nitrospira in the aquaponic system.
References
USEPA (2013) Aquatic Life Ambient Water Quality Criteria for Ammonia - Freshwater. Docket ID No EPA-HQ-OW-2009-0921
Hoseini SM, Vatnikov YA, Kulikov EV, Petrov AK, Hoseinifar SH, Van Doan H (2019) Effects of dietary arginine supplementation on ureagenesis and amino acid metabolism in common carp (Cyprinus carpio) exposed to ambient ammonia. Aquaculture 511:734209
Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63:153–182
Wongkiew S, Popp BN, Khanal SK (2018) Nitrogen recovery and nitrous oxide (N2O) emissions from aquaponic systems: influence of plant species and dissolved oxygen. Int Biodeter Biodegrad 134:117–126
Alonso A, Camargo J (2003) Short-term toxicity of ammonia, nitrite, and nitrate to the aquatic snail Potamopyrgus antipodarum (Hydrobiidae, Mollusca). Bull Environ Contam Toxicol 70(5):1006–1012
Stormer J, Jensen FB, Rankin JC (1996) Uptake of nitrite, nitrate, and bromide in rainbow trout, (Oncorhynchus mykiss): effects on ionic balance. Can J Fish Aquat Sci 53(9):1943–1950
Wongkiew S, Park M-R, Chandran K, Khanal SK (2018) Aquaponic systems for sustainable resource recovery: linking nitrogen transformations to microbial communities. Environ Sci Technol 52(21):12728–12739
Kuhn DD, Drahos DD, Marsh L, Flick GJ Jr (2010) Evaluation of nitrifying bacteria product to improve nitrification efficacy in recirculating aquaculture systems. Aquacult Eng 43(2):78–82
Foesel BU, Gieseke A, Schwermer C, Stief P, Koch L, Cytryn E, De La Torré JR, Van Rijn J, Minz D, Drake HL (2007) Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol Ecol 63(2):192–204
Tokuyama T, Mine A, Kamiyama K, Yabe R, Satoh K, Matsumoto H, Takahashi R, Itonaga K (2004) Nitrosomonas communis strain YNSRA, an ammonia-oxidizing bacterium, isolated from the reed rhizoplane in an aquaponics plant. J Biosci Bioeng 98(4):309–312
Itoi S, Niki A, Sugita H (2006) Changes in microbial communities associated with the conditioning of filter material in recirculating aquaculture systems of the pufferfish Takifugu rubripes. Aquaculture 256(1–4):287–295
Sugita H, Nakamura H, Shimada T (2005) Microbial communities associated with filter materials in recirculating aquaculture systems of freshwater fish. Aquaculture 243(1–4):403–409
Pedersen L-F, Pedersen PB, Nielsen JL, Nielsen PH (2009) Peracetic acid degradation and effects on nitrification in recirculating aquaculture systems. Aquaculture 296(3):246–254. https://doi.org/10.1016/j.aquaculture.2009.08.021
Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A (2015) Complete nitrification by Nitrospira bacteria. Nature 528(7583):504–509
van Kessel MA, Speth DR, Albertsen M, Nielsen PH, den Camp HJO, Kartal B, Jetten MS, Lücker S (2015) Complete nitrification by a single microorganism. Nature 528(7583):555–559
Fowler SJ, Palomo A, Dechesne A, Mines PD, Smets BF (2018) Comammox Nitrospira are abundant ammonia oxidizers in diverse groundwater-fed rapid sand filter communities. Environ Microbiol 20(3):1002–1015
Pinto AJ, Marcus DN, Ijaz UZ, Bautista-deloseSantos QM, Dick GJ, Raskin L (2016) Metagenomic evidence for the presence of comammox nitrospira-like bacteria in a drinking water system. Msphere 1(1):e00054-e15
Pjevac P, Schauberger C, Poghosyan L, Herbold CW, van Kessel MA, Daebeler A, Steinberger M, Jetten MS, Lücker S, Wagner M (2017) AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. Front Microbiol 8:1508
Bartelme RP, McLellan SL, Newton RJ (2017) Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing archaea and comammox Nitrospira. Front Microbiol 8:101
Schmautz Z, Graber A, Jaenicke S, Goesmann A, Junge R, Smits TH (2017) Microbial diversity in different compartments of an aquaponics system. Arch Microbiol 199(4):613–620
Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M (2001) In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol 67(11):5273–5284
Koch H, Lücker S, Albertsen M, Kitzinger K, Herbold C, Spieck E, Nielsen PH, Wagner M, Daims H (2015) Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. PNAS 112(36):11371–11376
Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M (2014) Development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS ONE 9(8):e105592
Dong X, Kleiner M, Sharp CE, Thorson E, Li C, Liu D, Strous M (2017) Fast and simple analysis of MiSeq amplicon sequencing data with MetaAmp. Front Microbiol 8:1461
Weinmaier T, Probst AJ, Duc MT, Ciobanu D, Cheng J-F, Ivanova N, Rattei T, Vaishampayan P (2015) A viability-linked metagenomic analysis of cleanroom environments: eukarya, prokaryotes, and viruses. Microbiome 3(1):62
Nurk S, Meleshko D, Korobeynikov A, Pevzner PA (2017) metaSPAdes: a new versatile metagenomic assembler. Genome Res 27(5):824–834
Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11(1):119
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12(1):59–60
Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH, Consortium U (2014) UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31(6):926–932
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8(3):275–282
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Wang Y, Ma L, Mao Y, Jiang X, Xia Y, Yu K, Li B, Zhang T (2017) Comammox in drinking water systems. Water Res 116:332–341
Palomo A, Pedersen AG, Fowler SJ, Dechesne A, Sicheritz-Pontén T, Smets BF (2018) Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J 12:1779–1793
Kim YM, Park D, Lee DS, Park JM (2007) Instability of biological nitrogen removal in a cokes wastewater treatment facility during summer. J Hazard Mater 141(1):27–32
Hochheimer JN, Wheaton F (1998) Biological filters: trickling and RBC design. In: Proc 2nd Intl Conf Recirculating Aquaculture 291–318
Pantanella E, Cardarelli M, Colla G, Rea E, Marcucci A (2012) Aquaponics vs. hydroponics: production and quality of lettuce crop. Acta Hortic 927:887–893. https://doi.org/10.17660/ActaHortic.2012.927.109
Hochmuth G (2001) Fertilizer management for greenhouse vegetables. Florida Greenhouse Vegetable Production Handbook 3:13–31
Rakocy JE, Masser MP, Losordo TM (2006) Recirculating aquaculture tank production systems: aquaponics—integrating fish and plant culture. SRAC Publication 454:1–16
Brown MN, Briones A, Diana J, Raskin L (2012) Ammonia-oxidizing archaea and nitrite-oxidizing nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS Microbiol Ecol 83(1):17–25
Costa E, Pérez J, Kreft J-U (2006) Why is metabolic labour divided in nitrification? Trends Microbiol 14(5):213–219. https://doi.org/10.1016/j.tim.2006.03.006
Acknowledgements
The authors thank the private operator of the aquaponics system for the permission to take samples and using the data for scientific work.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
JH and RM designed the study and the experiments. RM performed the sampling and JH did the analysis. JH, HM, and AP performed the bioinformatics. JH, HM, AP, and RM wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no financial or other conflicts. The aquaponic system was run by a private third party. This study only monitored the microbial communities in the system and analysed the chemical data with permission to use all data from the private aquaponics operator.
Ethical Approval
The authors are not responsible for the ethical treatment of animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Heise, J., Müller, H., Probst, A.J. et al. Ammonium Removal in Aquaponics Indicates Participation of Comammox Nitrospira. Curr Microbiol 78, 894–903 (2021). https://doi.org/10.1007/s00284-021-02358-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02358-3