Abstract
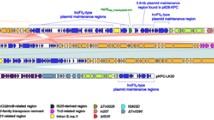
The IncX family of plasmids has recently been expanded to include at least four subtypes, IncX1–IncX4. The revised classification provides an opportunity for improving our understanding of the sequence diversity of the IncX plasmids and the resistance genes they carried. We described the complete nucleotide sequence of a novel IncX3 plasmid, pKPC-NY79 (42,447 bp) from a sequence-type 258 Klebsiella pneumoniae strain that was isolated from a patient who was hospitalized in New York, United States. In pKPC-NY79, the plasmid scaffold and genetic load region were highly similar to homologous regions in pIncX-SHV (IncX3, JN247852) and the bla KPC carrying pKpQIL (IncFIIk, GU595196), respectively, indicating that it has possibly arisen through recombination of plasmids. The bla KPC-2 gene, as part of a transposon Tn4401a, was found within the genetic load region. The backbone of pKPC-NY79 differs from pIncX-SHV by a deletion involving the gene tandem hns–topB (encoding H-NS protein and topoisomerase III, respectively) and a putative ATPase gene. Unexpectedly, the impact of the hns–topB deletion on host fitness and plasmid stability was found to be small. In conclusion, the findings contribute to a better understanding of the plasmid platforms carrying bla KPC and of variations in the backbone of the IncX3 plasmids.

Similar content being viewed by others
References
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ (2005) Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228
Castanheira M, Costello AJ, Deshpande LM, Jones RN (2012) Expansion of clonal complex 258 KPC-2-producing Klebsiella pneumoniae in Latin American hospitals: report of the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 56:1668–1669
Clinical and Laboratory Standards Institute (2012) Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement M100–S22. CLSI, Wayne
Cottell JL, Webber MA, Piddock LJ (2012) Persistence of transferable extended-spectrum-beta-lactamase resistance in the absence of antibiotic pressure. Antimicrob Agents Chemother 56:4703–4706
Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S (2005) Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182
Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ (2007) An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science 315:251–252
Garcia-Fernandez A, Villa L, Carta C, Venditti C, Giordano A, Venditti M, Mancini C, Carattoli A (2012) Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother 56:2143–2145
Gootz TD, Lescoe MK, Dib-Hajj F, Dougherty BA, He W, Della-Latta P, Huard RC (2009) Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob Agents Chemother 53:1998–2004
Ho PL, Li Z, Lai EL, Chiu SS, Cheng VC (2012) Emergence of NDM-1-producing Enterobacteriaceae in China. J Antimicrob Chemother 67:1553–1555
Ho PL, Li Z, Lo WU, Cheung YY, Lin CH, Sham PC, Cheng VC, Ng TK, Chow KH (2012) Identification and characterization of a novel incompatibility group X3 plasmid carrying bla NDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect 1:e39
Ho PL, Lo WU, Wong RC, Yeung MK, Chow KH, Que TL, Tong AH, Bao JY, Lok S, Wong SS (2011) Complete sequencing of the FII plasmid pHK01, encoding CTX-M-14, and molecular analysis of its variants among Escherichia coli from Hong Kong. J Antimicrob Chemother 66:752–756
Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AH, Bao JY, Lok S, Lo JY (2011) Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One 6:e17989
Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A (2012) Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid 68:43–50
Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, Nolan LK (2007) Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol 73:1976–1983
Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG (2009) Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370
Leavitt A, Chmelnitsky I, Carmeli Y, Navon-Venezia S (2010) Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother 54:4493–4496
Naas T, Cuzon G, Truong HV, Nordmann P (2012) Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother 56:4753–4759
Navon-Venezia S, Leavitt A, Schwaber MJ, Rasheed JK, Srinivasan A, Patel JB, Carmeli Y (2009) First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob Agents Chemother 53:818–820
Norman A, Hansen LH, She Q, Sorensen SJ (2008) Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59–74
Novais A, Canton R, Moreira R, Peixe L, Baquero F, Coque TM (2007) Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob Agents Chemother 51:796–799
Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L (2009) Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother 53:4333–4338
Sota M, Yano H, Hughes JM, Daughdrill GW, Abdo Z, Forney LJ, Top EM (2010) Shifts in the host range of a promiscuous plasmid through parallel evolution of its replication initiation protein. ISME J 4:1568–1580
Swinnen IA, Bernaerts K, Dens EJ, Geeraerd AH, Van Impe JF (2004) Predictive modelling of the microbial lag phase: a review. Int J Food Microbiol 94:137–159
Takeda T, Yun CS, Shintani M, Yamane H, Nojiri H (2011) Distribution of genes encoding nucleoid-associated protein homologs in plasmids. Int J Evol Biol 2011:685015
Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL (2012) Carbapenemases in Klebsiella pneumoniae and Other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707
Wolter DJ, Kurpiel PM, Woodford N, Palepou MF, Goering RV, Hanson ND (2009) Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob Agents Chemother 53:557–562
Acknowledgments
This work was supported by Grants from the Research Fund for the Control of Infectious Diseases (RFCID) of the Health and Food Bureau of the Government of the HKSAR. We thank Pak-Chung Sham, Levina Lam, Ben Ho, and Wilson Chan of the Centre for Genomic Science, University of Hong Kong for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ho, PL., Cheung, YY., Lo, WU. et al. Molecular Characterization of an Atypical IncX3 Plasmid pKPC-NY79 Carrying bla KPC-2 in a Klebsiella pneumoniae . Curr Microbiol 67, 493–498 (2013). https://doi.org/10.1007/s00284-013-0398-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-013-0398-2