Abstract
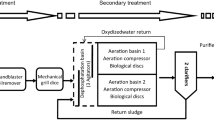
A pilot-scale membrane bioreactor (MBR) and a conventional activated sludge system (CAS) were in parallel operated to investigate the impact of the separation technology on the structure and functionality of the selected microbial community. Microbial communities as well as nitrogen removal efficiency of the biomass were characterized. Kinetics and microbial community structure turned out to be duly correlated. The impact of the separation technology on selective conditions and, in particular, the higher variability of solid separation efficiency in CAS with respect to MBR pilot plant possibly represented the main factor influencing the selection of bacterial communities. Concerning nitrifiers, bacteria of the genus Nitrospira were predominant in the MBR. This was in accordance with kinetics of nitrite-oxidizing bacteria that suggested the presence of k-strategists, while r-strategists were selected in the CAS plant, possibly because of the presence of transient higher concentrations of nitrite (in the range of 0.05–0.18 and of 0.05–4.4 mg \( {\text{NO}}_{2}^{ - } \)-N L−1 in the MBR and CAS effluents, respectively). An unexpectedly high presence of bacteria belonging to two specific phylogenetic clades of Planctomycetes was found in both reactors.






Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Baek SH, Pagilla K (2009) Microbial community structures in Conventional Activated Sludge System and Membrane Bioreactor (MBR). Biotechnol Bioprocess Eng 14:848–853
Calderon K, Rodelas B, Cabirol N, Gonzalez-Lopez J, Noyola A (2011) Analysis of microbial communities developed on the fouling layers of a membrane-coupled anaerobic bioreactor applied to wastewater treatment. Bioresour Technol 102:4618–4627
Calderon K, Gonzalez-Martinez A, Montero-Puente C, Reboleiro-Rivas P, Poyatos JM, Juarez-Jimenez B, Martinez-Toledo M, Rodelas B (2012) Bacterial community structure and enzyme activities in a membrane bioreactor (MBR) using pure oxygen as an aeration source. Bioresour Technol 103:87–94
Cases V, Alonso V, Argandoña V, Rodriguez M, Prats D (2011) Endocrine disrupting compounds: a comparison of removal between conventional activated sludge and membrane bioreactors. Desalination 272(1–3):240–245
Chouari R, Le Paslier D, Daegelen P, Ginestet P, Weissenbach J, Sghir A (2003) Molecular evidence for novel planctomycete diversity in a municipal wastewater treatment plant. Appl Environ Microbiol 69(12):7354–7363
Clara M, Strenna B, Gans O, Martinez E, Kreuzingera N, Kroissa H (2005) Removal of selected pharmaceuticals, fragrances and endocrine disrupting compounds in a membrane bioreactor and conventional wastewater treatment plants. Water Res 39:4797–4807
Curtis TP, Head IM, Graham DW (2003) Theoretical ecology for engineering biology. Environ Sci Technol 37:64A–70A
Daebel H, Manser R, Gujer W (2007) Exploring temporal variations of oxygen saturation constants of nitrifying bacteria. Water Res 41(5):1094–1102
Daims H, Brühl A, Amann R, Schleifer KH, Wagner M (1999) The domain specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444
Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M (2001) In situ characterization of Nitrospira-like Nitrite-Oxidizing bacteria active in waste water treatment plants. Appl Environ Microbiol 67:5273–5284
De Wever H, Weiss S, Reemtsma T, Vereecken J, Müller J, Knepper T, Rörden O, Gonzalez S, Barcelo D, Hernando MD (2007) Comparison of sulfonated and other micropollutants removal in membrane bioreactor and conventional wastewater treatment. Water Res 41(4):935–945
Drews A (2010) Membrane fouling in membrane bioreactors: characterisation, contradictions, cause and cures. J Membr Sci 363:1–28
Dytczak MA, Londry KL, Oleszkiewicz JA (2008) Activated sludge operational regime has significant impact on the type of nitrifying community and its nitrification rates. Water Res 42(8–9):2320–2328
Fenu A, Guglielmi G, Jimenez J, Spèrandio M, Saroj D, Lesjean B, Brepols C, Thoeye C, Nopens I (2010) Activated sludge model (ASM) based modelling of membrane bioreactor (MBR) processes: a critical review with special regard to MBR specificities. Water Res 44(15):4272–4294
Fenu A, Roels J, Wambecq T, De Gussem K, Thoeye C, De Gueldre G, Van De Steene B (2010) Energy audit of a full scale MBR system. Desalination 262(1–3):121–128
Gil JA, Túa L, Rueda A, Montaño B, Rodríguez M, Prats D (2010) Monitoring and analysis of the energy cost of an MBR. Desalination 250(3):997–1001
Gonzalez S, Petrovic M, Barceló D (2007) Removal of a broad range of surfactants from municipal wastewater—comparison between membrane bioreactor and conventional activated sludge treatment. Chemosphere 67:335–343
Griepenburg U, Ward-Rainey N, Mohamed S, Schlesner H, Marxsen H, Rainey FA, Stackebrandt E, Auling G (1999) Phylogenetic diversity, polyamine pattern and DNA base composition of members of the order Planctomycetales. Int J Syst Bacteriol 49:689–696
Hugenholtz P, Huber T (2003) Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int J Syst Evol Microbiol 53:289–293
Kim DJ, Kim SH (2006) Effect of nitrite concentration on the distribution and competition of nitrite-oxidizing bacteria in nitratation reactor systems and their kinetic characteristic. Water Res 40:887–894
Koops HP, Purkhold U, Pommerening-Roser A, Timmermann G, Wagner M (2006) The lithoautotrophic ammonia-oxidizing bacteria. In: The prokaryotes: a handbook on the biology of bacteria, 3rd edn. Springer, New York, pp 778–811
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–147
Li HQ, Jiku F, Schroder HF (2000) Assessment of the pollutant elimination efficiency by gas chromatography/mass spectrometry, liquid chromatography–mass spectrometry and -tandem mass spectrometry—comparison of conventional and membrane-assisted biological wastewater treatment processes. J Chromatogr 889:155–176
Loy A, Daims H, Wagner M (2002) Activated sludge: molecular techniques for determining community composition. In: Bitton G (ed) The encyclopedia of environmental microbiology. Wiley, New York, pp 26–43
Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, Schleifer KH, Wagner M (2002) Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol 68:5064–5081
Loy A, Maixner F, Wagner M, Horn M (2007) ProbeBase—an online resource for rRNA-targeted oligonucleotide probes: new features. Nucleic Acids Res 35:D800–D804
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lüßmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Magurran AE (1998) Ecological diversity and its measurement. Chapman and Hall, London
Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, Daims H (2006) Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol 8(8):1487–1495
Manser R, Gujer W, Siegrist H (2005) Consequences of mass transfer effects on the kinetics of nitrifiers. Water Res 39(19):4633–4642
Manser R, Gujer W, Siegrist H (2006) Decay processes of nitrifying bacteria in biological wastewater treatment systems. Water Res 40(12):2416–2426
Manz W, Amann R, Ludwig W, Wagner M, Schleifer KH (1992) Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol 15:593–600
Morgan-Sagastume F, Larsen P, Nielsen JL, Nielsen PH (2008) Characterization of the loosely attached fraction of activated sludge bacteria. Water Res 42(4–5):843–854
Munz G, Mori G, Salvadori L, Barberio C, Lubello C (2008) Process efficiency and microbial monitoring in MBR (membrane bioreactor) and CASP (conventional activated sludge process) treatment of tannery wastewater. Bioresour Technol 99:8559–8564
Munz G, Lubello C, Oleszkiewicz JA (2011) Modeling the decay of ammonium oxidizing bacteria. Water Res 45(2):557–564
Munz G, Szoke N, Oleszkiewicz JA (2012) Effect of ammonia oxidizing bacteria (AOB) kinetics on bioaugmentation. Bioresour Technol 125:88–96
Neef A, Amann R, Schlesner H, Schleifer KH (1998) Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257–3266
Parco V, du Toit G, Wentzel M, Ekama G (2007) Biological nutrient removal in membrane bioreactors: denitrification and phosphorus removal kinetics. Water Sci Technol 56(6):125–134
Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J, Glockner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Rivière D, Desvignes V, Pelletier E, Chaussonnerie S, Guermazi S, Weissenbach J, Li T, Camacho P, Sghir A (2009) Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J 3:700–714
Sahar E, Ernst M, Godehardt M, Hein A, Herr J, Kazner C, Melin T, Cikurel H, Aharoni A, Messalem R, Brenner A, Jekel M (2011) Comparison of two treatments for the removal of selected organic micropollutants and bulk organic matter: conventional activated sludge followed by ultrafiltration versus membrane bioreactor. Water Sci Technol 63(4):733–740
Satoh H, Yamakawa T, Kindaichi T, Ito T, Okabe S (2006) Community structures and activities of nitrifying and denitrifying bacteria in industrial wastewater-treating biofilms. Biotechnol Bioeng 94:762–772
Schmid MC, Maas B, Dapena A, van de Pas-Schoonen K, van de Vossenberg J, Kartal B, van Niftrik L, Schmidt I, Cirpus I, Kuenen JC, Wagner M, Sinninghe Damsté JS, Kuypers M, Revsbech NP, Mendez R, Jetten MS, Strous M (2005) Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl Environ Microbiol 71:1677–1684
Silva CC, Jesus EC, Torres AP, Sousa MP, Santiago VM, Oliveira VM (2010) Investigation of bacterial diversity in membrane bioreactor and conventional activated sludge processes from petroleum refineries using phylogenetic and statistical approaches. J Microbiol Biotechnol 20(3):447–459
Stephen JR, Kowalchuk GA, Bruns MAV, McCaig AE, Phillips CJ, Embley TM, Prosser JI (1998) Analysis of beta-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl Environ Microbiol 64:2958–2965
Tindall BJ, Rossellò-Mòra R, Busse HJ, Ludwig W, Kampfer P (2010) Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60:249–266
Valderrama C, Ribera G, Bahí N, Rovira M, Giménez T, Nomen R, Lluch S, Yuste M, Martinez-Lladó X (2012) Winery wastewater treatment for water reuse purpose: conventional activated sludge versus membrane bioreactor (MBR): a comparative case study. Desalination 306:1–7
Wang X, Wen X, Yan H, Ding K, Zhao F, Hu M (2011) Bacterial community dynamics in a functionally stable pilot-scale wastewater treatment plant. Bioresour Technol 102(3):2352–2357
Xia S, Li J, He S, Xie K, Wang X, Zhang Y, Duan L, Zhang Z (2010) The effect of organic loading on bacterial community composition of membrane biofilms in a submerged polyvinyl chloride membrane bioreactor. Bioresour Technol 101(17):6601–6609
Acknowledgments
Authors wish to thank T. Bhuiyan and G. Aulicino for their help in laboratory procedures. S. Gabrielli′s help with graphic artwork is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chiellini, C., Munz, G., Petroni, G. et al. Characterization and Comparison of Bacterial Communities Selected in Conventional Activated Sludge and Membrane Bioreactor Pilot Plants: A Focus on Nitrospira and Planctomycetes Bacterial Phyla . Curr Microbiol 67, 77–90 (2013). https://doi.org/10.1007/s00284-013-0333-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-013-0333-6