Abstract
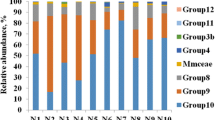
In order to assess methanogen diversity in feces of pigs, archaeal 16S rRNA gene clone libraries were constructed from feces of the pig. After the amplification by PCR using primers Met86F and Met1340R, equal quantities of PCR products from each of the five pigs were mixed together and used to construct the library. Sequence analysis showed that the 74 clones were divided into ten phylotypes as defined by RFLP analysis. Phylogenetic analysis showed that three phylotypes were most closely affiliated with the genus Methanobrevibacter (46% of clones). The library comprised 55.4% unidentified euryarchaeal clones. Three phylotypes (LMG4, LMG6, LMG8) were not closely related to any known Euryarchaeota sequences. The phylogenetic analysis indicated that the archaea found in the libraries were all clustered into the Euryarchaeota. The data from the phylogenetic tree showed that those sequences belonged to three monophyletic groups. Phylotypes LGM2 and LGM7 grouped within the genus Methanobrevibacter. Phylotypes LGM4, LGM6, LGM8 and LGM9 grouped within the genus Methanosphaera. Other phylotypes grouped together, and formed a distantly related sister group to Aciduliprofundum boonei and species of the Thermoplasmatales including Thermoplasma volcanium and Thermoplasm acidophilum. Our results showed that methanogens belonging to the genus Methanobrevibacter were predominant in pig feces, and that many unique unknown archaea sequences were also found in the library. Nevertheless, whether these unique sequences represent new taxonomic groups and their role in the pig gut need further investigation.

Similar content being viewed by others
References
Bergey DH (1994) The methanogens. In: Holt JG (ed) Bergey’s manual of determinative bacteriology, 9th edn. Williams and Wilkins Co, Baltimore, pp 719–725
Miller TL, Wolin MJ (1986) Methanogens in human and animal intestinal tracts. Syst Appl Microbiol 7:223–229
Miller TL, Wolin MJ, Kusel EA (1986) Isolation and characterization of methanogens from animal feces. Syst Appl Microbiol 8:234–238
Monteny GJ, Groenestein CM, Hilhorst MA (2001) Interactions and coupling between emissions of methane and nitrous oxide from animal husbandry. Nutr Cycl Agroecosys 60:123–132
Pei C-X, Mao S-Y, Cheng Y-F, Zhu W-Y (2010) Diversity, abundance and novel 16S rRNA gene sequences of methanogens in rumen liquid, solid and epithelium fractions of Jinnan cattle. Animal 4(1):20–29
Whitford MF, Teather RM, Forster RJ (2001) Phylogenetic analysis of methanogens from the bovine rumen. BMC Microbiol 1:1–5
Woese CR, Kandler O, Wheelis ML (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. PANS 87:4576–4579
Wright ADG, Auckland CH, Lynn DH (2007) Molecular diversity of methanogens in feedlot cattle from Ontario and Prince Edward Island, Canada. Appl Environ Microbiol 73:4206–4210
Wright ADG, Pimm C (2003) Improved strategy for presumptive identification of methanogens using 16S riboprinting. J Microbiol Methods 55:337–349
Wright ADG, Toovey AF, Pimm CL (2006) Molecular identification of methanogenic archaea from sheep in Queensland, Australia reveals more uncultured novel archaea. Anaerobe 12:134–139
Wright ADG, Williams AJ, Winder B et al (2004) Molecular diversity of rumen methanogens from sheep in Western Australia. Appl Environ Microbiol 70:1263–1270
Zoetendal EG, Akkermans AD, De Vos WM (1998) Temperature gradient gel electrophoresis analysis from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol 64:3854–3859
Acknowledgments
This work was supported by Natural Science Foundation of China (NSFC) (30810103909), R&D Special Fund for Public Welfare In Agriculture of China (200903003).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mao, SY., Yang, CF. & Zhu, WY. Phylogenetic Analysis of Methanogens in the Pig Feces. Curr Microbiol 62, 1386–1389 (2011). https://doi.org/10.1007/s00284-011-9873-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-011-9873-9