Abstract
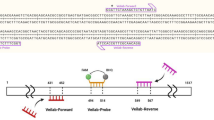
The relationship between copy numbers of internal transcribed spacer 1 (ITS1) and biomass or zoospore count of anaerobic fungi was studied to develop a quantitative real-time PCR-based monitoring method for fungal biomass or population in the rumen. Nine fungal strains were used to determine the relationship between ITS1 copy number and fungal biomass. Rumen fluid from three sheep and a cow were used to determine the relationship between ITS1 copy number and fungal population. ITS1 copy number was determined by real-time PCR with a specific primer set for anaerobic fungi. Freeze-dried fungal cells were weighed for fungal biomass. Zoospore counts were determined by the roll-tube method. A positive correlation was observed between both ITS1 copy number and dry weight and ITS1 copy number and zoospore counts, suggesting that the use of ITS1 copy numbers is effective for estimating fungal biomass and population density. On the basis of ITS1 copy numbers, fluctuations in the fungal population in sheep rumen showed that although the values varied among individual animals, the fungal population tended to decrease after feeding. In the present study, a culture-independent method was established that will provide a powerful tool for understanding the ecology of anaerobic fungi in the rumen.

Similar content being viewed by others
References
Brookman JL, Mennim G, Trinci APJ et al (2000) Identification and characterization of anaerobic gut fungi using molecular methodologies based on ribosomal ITS 1 and 18S rRNA. Microbiology 146:393–403
Dehority BA, Orpin CG (1997) Development of, and natural fluctuations in, rumen microbial populations. In: Hobson PN, Stewart CS (eds) The rumen microbial ecosystem. Elsevier, London, pp 196–245
Denman SE, McSweeney CS (2006) Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol 58:572–582
Joblin KN (1981) Isolation, enumeration, and maintenance of rumen anaerobic fungi in roll tubes. Appl Environ Microbiol 42:1119–1122
Li J, Heath IB (1992) The phylogenetic relationships of the anaerobic Chytridiomycetous gut fungi (Neocallimasticaceae and the Chytriomycota. 1. Cladistic analysis of ribosomal RNA sequences. Can J Bot 70:1738–1746
Matsui H, Ban-Tokuda T (2008) Studies on carboxymethyl cellulase and xylanase activities of an anaerobic fungal isolate CR4 from the bovine rumen. Curr Microbiol 57:615–619
Matsui H, Ushida K, Kojima Y (1997) Effect of dietary concentrate on fungal zoosporogenesis in sheep rumen. Asian-Australas J Anim Sci 10:599–602
Orpin CG (1975) Studies on the rumen flagellate Neocallimastix frontalis. J Gen Microbiol 91:249–262
Orpin CG (1994) Anaerobic fungi: taxonomy, biology, and distribution in nature. In: Mountfort DO, Orpin CG (eds) Anaerobic fungi. Marcel Dekker, Inc., New York, pp 1–45
Orpin CG, Joblin KN (1997) Anaerobic fungi. In: Hobson PN, Stewart CS (eds) The rumen microbiol ecosystem, 2nd edn. Blackie Academic and Professional, Chapman and Hall, pp 129–150
Ozutsumi Y, Tajima K, Takenaka A et al (2006) Real-time PCR detection of the effects of protozoa on rumen bacteria in cattle. Curr Microbiol 52:158–162
Sekhavati MH, Mesgaran MD, Nassiri MR et al (2009) Development and use of quantitative competitive PCR assays for relative quantifying rumen anaerobic fungal populations in both in vitro and in vivo systems. Mycol Res 113:1146–1153
Shinkai T, Kobayashi Y (2007) Localization of ruminal cellulolytic bacteria on plant fibrous materials as determined by fluorescence in situ hybridization and real-time PCR. Appl Environ Microbiol 73:1646–1652
Tajima K, Aminov RI, Nagamine T et al (2001) Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl Environ Microbiol 67:2766–2774
Trinci APJ, Davies DR, Gull K et al (1994) Anaerobic fungi in herbivorous animals. Mycol Res 98:129–152
Uyeno Y, Sekiguchi Y, Tajima K et al (2010) An rRNA-based analysis for evaluating the effect of heat stress on the rumen microbial composition of Holstein heifers. Anaerobe 16:27–33
Williams AG, Orpin CG (1987) Polysaccharide-degrading enzymes formed by three species of anaerobic fungi grown on a range of carbohydrate substrates. Can J Microbiol 33:418–426
Williams YJ, Popovski S, Rea SM et al (2009) A vaccine against rumen methanogens can alter the composition of archaeal populations. Appl Environ Microbiol 75:1860–1866
Wood TM, Wilson CA, McCrae SI et al (1986) A highly active extracellular cellulase from the anaerobic rumen fungus Neocallimastix frontalis. FEMS Microbiol Lett 34:37–40
Wubah DA, Fuller MS, Akin DE (1991) Studies on Caecomyces communis: morphology and development. Mycologia 83:303–310
Zoetendal EG, Collier CT, Koike S et al (2004) Molecular ecological analysis of the gastrointestinal microbiota: a review. J Nutr 134:465–472
Acknowledgments
The authors would like to thank Dr. Yasunari Yamamoto (Mie Prefecture Livestock Research Institute, Mie Prefecture, Japan) for providing rumen fluid and Professor Kazuo Sakka (Graduate School of Mie University, Japan) for his instruction on real-time PCR. This study was supported by the Integrated Research for Developing Japanese-style Forage Feeding System to Increase Forage Self-Support Ratio of the Ministry of Agriculture, Forestry, and Fisheries of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lwin, K.O., Hayakawa, M., Ban-Tokuda, T. et al. Real-Time PCR Assays for Monitoring Anaerobic Fungal Biomass and Population Size in the Rumen. Curr Microbiol 62, 1147–1151 (2011). https://doi.org/10.1007/s00284-010-9843-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-010-9843-7