Abstract
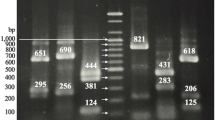
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis of amplified DNA fragment of the 16S and 23S rRNA genes was performed on 35 Helicobacter, 24 Campylobacter, and 15 Arcobacter strains. PCR amplification generated a 1004-bp fragment of 16S rDNA and a 2.6-Kbp fragment of 23S rDNA from each strain. The amplicons were digested with DdeI and HpaII, respectively. For both assays, distinctive profiles were obtained for each genus. 23S rDNA PCR-RFLP analysis with HpaII enzyme identified Campylobacter and Helicobacter strains at the species level. Analysis of 16S rRNA gene with DdeI enzyme was not useful for the specific identification of Campylobacter and Arcobacter, although it discriminated among Helicobacter species. The PCR-RFLP technique allowed for the discrimination among these three related genus with only one restriction enzyme; therefore it can be a simple, rapid, and useful method for routine identification.


Similar content being viewed by others
Literature Cited
Atabay HI, Corry JEL, On SLW (1998) Diversity and prevalence of Arcobacter spp. in broiler chickens. J Appl Microbiol 84:1007–1016
Bastyns K, Cartuyvels D, Chapelle S, et al. (1995) A variable 23S rDNA region is a useful discriminating target for genus-specific and species-specific PCR amplification in Arcobacter species. Syst Appl Microbiol 18:353–356
Cardarelli-Leite P, Blom K, Patton CM, et al. (1996) Rapid identification of Campylobacter species by restriction fragment length polymorphism analysis of a PCR-amplified fragment of the gene coding for 16S rRNA. J Clin Microbiol 34:62–67
Choi YK, Han JH, Joo HS (2001) Identification of novel Helicobacter species in pig stomachs by PCR and partial sequencing. J Clin Microbiol 39:3311–3315
Cover TL, Blaser MJ (1995) Helicobacter pylori: A bacterial cause of gastritis, peptic ulcer disease, and gastric cancer. ASM Features 61:21–26
de Boer WA, Tytgat GN (2000) Regular review: Treatment of Helicobacter pylori infection. Br Med J 320:31–34
Denis M, Soumet C, Revoal K, et al. (1999) Development of a m-PCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett Appl Microbiol 29:406–410
Engvall EO, Brandstrom B, Gunnarsson A, et al. (2002) Validation of a polymerase chain reaction/restriction enzyme analysis method for species identification of thermophilic campylobacters isolated from domestic and wild animals. J Appl Microbiol 92:47–54
Fermér C, Engvall EO (1999) Specific PCR identification and differentiation of the thermophilic campylobacters, Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J Clin Microbiol 37:3370–3373
Fox JG (2002) The non-H pylori helicobacters: Their expanding role in gastrointestinal and systemic diseases. Gut 50:273–283
Fox JG, Dewhirst FE, Shen Z, et al. (1998) Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterol 114:755–763
Frost JA (2001) Current epidemiological issues in human campylobacteriosis. Symp Ser Soc Appl Microbiol 30:85S–95S
González I, Grant KA, Richardson PT, et al. (1997) Specific identification of the enteropathogens Campylobacter jejuni and Campylobacter coli by using a PCR test based on the ceuE gene encoding a putative virulence determinant. J Clin Microbiol 35:759–763
Hsueh PR, Teng LJ, Hung CC, et al. (1999) Septic shock due to Helicobacter fennelliae in a non-human immunodeficiency virus-infected heterosexual patient. J Clin Microbiol 37:2084–2086
Hurtado A, Owen RJ (1997a) A rapid identification scheme for Helicobacter pylori and other species of Helicobacter based on 23S rRNA gene polymorphisms. Syst Appl Microbiol 20:222–231
Hurtado A, Owen RJ (1997b) A molecular scheme based on 23S rRNA gene polymorphisms for rapid identification of Campylobacter and Arcobacter species. J Clin Microbiol 35:2401–2404
Huysmans MB, Turnidge JD, Williams JH (1995) Evaluation of API Campy in comparison with conventional methods for identification of thermophilic campylobacters. J Clin Microbiol 33:3345–3346
International Agency for Research on Cancer (1994) Schistosomotes, liver flukes and Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum 61:1–241
Jalava K, Hielm S, Hirvi U, Hanninen ML (1999) Evaluation of a molecular identification scheme based on 23S rRNA gene polymorphisms for differentiating canine and feline gastric Helicobacter spp. Lett Appl Microbiol 28:269–274
Jauk V, Neubauer C, Szolgyenyi W, Vasicek L (2003) Phenotypic and genotypic differentiation of Campylobacter spp. isolated from Austrian broiler farms: A comparison. Avian Pathol 32:33–37
Kabeya H, Kobayashi Y, Maruyama S, Mikami T (2003) One-step polymerase chain reaction-based typing of Arcobacter species. Int J Food Microbiol 81:163–168
Linton D, Lawson AJ, Owen RJ, Stanley J (1997) PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J Clin Microbiol 35:2568–2572
Mansfield LP, Forshythe SJ (2000) Arcobacter butzleri, A. skirrowii and A. cryaerophilus-potencial emerging human pathogens. Rev Med Microbiol 11:161–170
Marshall SM, Melito PL, Woodward DL, et al. (1999) Rapid identification of Campylobacter, Arcobacter, and Helicobacter isolates by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene. J Clin Microbiol 37:4158–4160
Melito PL, Woodward DL, Bernard KA, et al. (2000) Differentiation of clinical Helicobacter pullorum isolates from related Helicobacter and Campylobacter species. Helicobacter 5:142–147
Moreno Y, Ferrús MA, Vanoostende A, et al. (2002) Comparison of 23S polymerase chain reaction and amplified fragment length polymorphism techniques as typing systems for thermophilic campylobacters. FEMS Microbiol Lett 211:97–103
On SLW (2001) Taxonomy of Campylobacter, Arcobacter, Helicobacter and related bacteria: Current status, future prospects and immediate concerns. J Appl Microbiol 90:1S–15S
On SLW (1996) Identification methods for campylobacters, helicobacters and related organisms. Clin Microbiol Rev 9:405–422
O’Rourke JL, Grehan M, Lee A (2001) Non-pylori helicobacter species in humans. Gut 49:601–606
Pearson AD, Greenwood MH, Feltham RK, et al. (1996) Microbial ecology of Campylobacter jejuni in a United Kingdom chicken supply chain: Intermittent common source, vertical transmission, and amplification by flock propagation. Appl Environ Microbiol 62:4614–4620
Rautelin H, Jusufovic J, Hänninen ML (1999) Identification of hippurate-negative thermophilic campylobacters. Diagn Microbiol Infect Dis 35:9–12
Shen Z, Fox JG, Dewhirst FE, et al. (1997) Helicobacter rodentium sp. nov., a urease-negative Helicobacter species isolated from laboratory mice. Int J Syst Bacteriol 47:627–634
Solnick JV, Schauer DB (2001) Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin Microbiol Rev 14:59–97
Steinhauserova I, Ceskova J, Fojtikova K, Obrovska I (2001) Identification of thermophilic Campylobacter spp. by phenotypic and molecular methods. J Appl Microbiol 90:470–475
Stucki U, Frey J, Nicolet J, Burnens AP (1995) Identification of Campylobacter jejuni on the basis of a species-specific gene that encodes a membrane protein. J Clin Microbiol 33:855–859
Vandamme P, Falsen E, Rossau R, et al. (1991) Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: Emendation of generic descriptions and proposal of Arcobacter gen. nov. Int J Syst Bacteriol 41:88–103
Weir SC, Gibert CL, Gordin FM, et al. (1999) An uncommon Helicobacter isolate from blood: Evidence of a group of Helicobacter spp. pathogenic in AIDS patients. J Clin Microbiol 37:2729–2733
Wesley IV (1997) Helicobacter and Arcobacter: Potential human foodborne pathogens? Trends Food Sci 89:293–299
Wilson K (1987) Preparation of genomic DNA from bacteria. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, et al. (eds) Current protocols in molecular biology, unit 2.4.1. Wiley, New York, NY, pp 241–242
Acknowledgments
This work was supported by Research Project AGL2002-04480-C03-03 from Ministerio de Ciencia y Tecnología, Spain (National and FEDER fundings). A. González is the recipient of a Predoctoral FPU grant from Ministerio de Educación y Ciencia, Spain (AP2000-0795).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
González, A., Moreno, Y., González, R. et al. Development of a Simple and Rapid Method Based on Polymerase Chain Reaction–Based Restriction Fragment Length Polymorphism Analysis to Differentiate Helicobacter, Campylobacter, and Arcobacter Species. Curr Microbiol 53, 416–421 (2006). https://doi.org/10.1007/s00284-006-0168-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-006-0168-5