Abstract
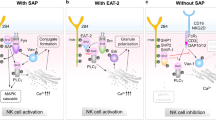
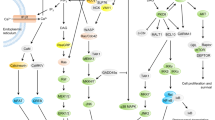
One or more of the signaling lymphocytic activation molecule (SLAM) family (SLAMF) of cell surface receptors, which consists of nine transmembrane proteins, i.e., SLAMF1-9, are expressed on most hematopoietic cells. While most SLAMF receptors serve as self-ligands, SLAMF2 and SLAMF4 use each other as counter structures. Six of the receptors carry one or more copies of a unique intracellular tyrosine-based switch motif, which has high affinity for the single SH2-domain signaling molecules SLAM-associated protein and EAT-2. Whereas SLAMF receptors are costimulatory molecules on the surface of CD4+, CD8+, and natural killer (NK) T cells, they also involved in early phases of lineage commitment during hematopoiesis. SLAMF receptors regulate T lymphocyte development and function and modulate lytic activity, cytokine production, and major histocompatibility complex-independent cell inhibition of NK cells. Furthermore, they modulate B cell activation and memory generation, neutrophil, dendritic cell, macrophage and eosinophil function, and platelet aggregation. In this review, we will discuss the role of SLAM receptors and their adapters in T cell function, and we will examine the role of these receptors and their adapters in X-linked lymphoproliferative disease and their contribution to disease susceptibility in systemic lupus erythematosus.




Similar content being viewed by others
References
Wang N, Morra M, Wu C, Gullo C, Howie D, Coyle T, Engel P, Terhorst C (2001) CD150 is a member of a family of genes that encode glycoproteins on the surface of hematopoietic cells. Immunogenetics 53:382–394
Tovar V, del Valle J, Zapater N, Martin M, Romero X, Pizcueta P, Bosch J, Terhorst C, Engel P (2002) Mouse novel Ly9: a new member of the expanding CD150 (SLAM) family of leukocyte cell-surface receptors. Immunogenetics 54:394–402
Fraser CC, Howie D, Morra M, Qiu Y, Murphy C, Shen Q, Gutierrez-Ramos JC, Coyle A, Kingsbury GA, Terhorst C (2002) Identification and characterization of SF2000 and SF2001, two new members of the immune receptor SLAM/CD2 family. Immunogenetics 53:843–850
Staunton DE, Thorley-Lawson DA (1987) Molecular cloning of the lymphocyte activation marker Blast-1. EMBO J 6:3695–3701
Yokoyama S, Staunton D, Fisher R, Amiot M, Fortin JJ, Thorley-Lawson DA (1991) Expression of the Blast-1 activation/adhesion molecule and its identification as CD48. J Immunol 146:2192–2200
Calpe S, Wang N, Romero X, Berger SB, Lanyi A, Engel P, Terhorst C (2008) The SLAM and SAP gene families control innate and adaptive immune responses. Adv Immunol 97:177–250
Engel P, Eck MJ, Terhorst C (2003) The SAP and SLAM families in immune responses and X-linked lymphoproliferative disease. Nat Rev Immunol 3:813–821
Cao E, Ramagopal UA, Fedorov A, Fedorov E, Yan Q, Lary JW, Cole JL, Nathenson SG, Almo SC (2006) NTB-A receptor crystal structure: insights into homophilic interactions in the signaling lymphocytic activation molecule receptor family. Immunity 25:559–570
Yan Q, Malashkevich VN, Fedorov A, Fedorov E, Cao E, Lary JW, Cole JL, Nathenson SG, Almo SC (2007) Structure of CD84 provides insight into SLAM family function. Proc Natl Acad Sci 104:10583–10588
Roda-Navarro P, Mittelbrunn M, Ortega M, Howie D, Terhorst C, Sanchez-Madrid F, Fernandez-Ruiz E (2004) Dynamic redistribution of the activating 2B4/SAP complex at the cytotoxic NK cell immune synapse. J Immunol 173:3640–3646
Howie D, Okamoto S, Rietdijk S, Clarke K, Wang N, Gullo C, Bruggeman JP, Manning S, Coyle AJ, Greenfield E, Kuchroo V, Terhorst C (2002) The role of SAP in murine CD150 (SLAM)-mediated T-cell proliferation and interferon gamma production. Blood 100:2899–2907
Rethi B, Gogolak P, Szatmari I, Veres A, Erdos E, Nagy L, Rajnavolgyi E, Terhorst C, Lanyi A (2006) SLAM/SLAM interactions inhibit CD40-induced production of inflammatory cytokines in monocyte-derived dendritic cells. Blood 107:2821–2829
Martin M, Del Valle JM, Saborit I, Engel P (2005) Identification of Grb2 as a novel binding partner of the signaling lymphocytic activation molecule-associated protein binding receptor CD229. J Immunol 174:5977–5986
Del Valle JM, Engel P, Martin M (2003) The cell surface expression of SAP-binding receptor CD229 is regulated via its interaction with clathrin-associated adaptor complex 2 (AP-2). J Biol Chem 278:17430–17437
Purtilo DT, Cassel CK, Yang JP, Harper R (1975) X-linked recessive progressive combined variable immunodeficiency (Duncan’s disease). Lancet 1:935–940
Harrington DS, Weisenburger DD, Purtilo DT (1987) Malignant lymphoma in the X-linked lymphoproliferative syndrome. Cancer 59:1419–1429
Egeler RM, de Kraker J, Slater R, Purtilo DT (1992) Documentation of Burkitt lymphoma with t(8;14) (q24;q32) in X-linked lymphoproliferative disease. Cancer 70:683–687
Schuster V, Terhorst C (2006) Primary immunodeficiency diseases: a molecular and cellular approach, 2nd edn. Oxford University Press, Oxford, pp 470–484
Morra M, Howie D, Grande MS, Sayos J, Wang N, Wu C, Engel P, Terhorst C (2001) X-linked lymphoproliferative disease: a progressive immunodeficiency. Annu Rev Immunol 19:657–682
Sumegi J, Huang D, Lanyi A, Davis JD, Seemayer TA, Maeda A, Klein G, Seri M, Wakiguchi H, Purtilo DT, Gross TG (2000) Correlation of mutations of the SH2D1A gene and Epstein–Barr virus infection with clinical phenotype and outcome in X-linked lymphoproliferative disease. Blood 96:3118–3125
Poy F, Yaffe MB, Sayos J, Saxena K, Morra M, Sumegi J, Cantley LC, Terhorst C, Eck MJ (1999) Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition. Mol Cell 4:555–561
Morra M, Silander O, Calpe S, Choi M, Oettgen H, Myers L, Etzioni A, Buckley R, Terhorst C (2001) Alterations of the X-linked lymphoproliferative disease gene SH2D1A in common variable immunodeficiency syndrome. Blood 98:1321–1325
Parolini O, Kagerbauer B, Simonitsch-Klupp I, Ambros P, Jaeger U, Mann G, Haas OA, Morra M, Gadner H, Terhorst C, Knapp W, Holter W (2002) Analysis of SH2D1A mutations in patients with severe Epstein–Barr virus infections, Burkitt’s lymphoma, and Hodgkin’s lymphoma. Ann Hematol 81:441–447
Rigaud S, Fondaneche MC, Lambert N, Pasquier B, Mateo V, Soulas P, Galicier L, Le Deist F, Rieux-Laucat F, Revy P, Fischer A, de Saint BG, Latour S (2006) XIAP deficiency in humans causes an X-linked lymphoproliferative syndrome. Nature 444:110–114
Latour S (2007) Natural killer T cells and X-linked lymphoproliferative syndrome. Curr Opin Allergy Clin Immunol 7:510–514
Marodi L, Notarangelo LD (2007) Immunological and genetic bases of new primary immunodeficiencies. Nat Rev Immunol 7:851–861
Rumble JM, Oetjen KA, Stein PL, Schwartzberg PL, Moore BB, Duckett CS (2009) Phenotypic differences between mice deficient in XIAP and SAP, two factors targeted in X-linked lymphoproliferative syndrome (XLP). Cell Immunol 259:82–89
Morra M, Simarro-Grande M, Martin M, Chen AS, Lanyi A, Silander O, Calpe S, Davis J, Pawson T, Eck MJ, Sumegi J, Engel P, Li SC, Terhorst C (2001) Characterization of SH2D1A missense mutations identified in X-linked lymphoproliferative disease patients. J Biol Chem 276:36809–36816
Wu C, Sayos J, Wang N, Howie D, Coyle A, Terhorst C (2000) Genomic organization and characterization of mouse SAP, the gene that is altered in X-linked lymphoproliferative disease. Immunogenetics 51:805–815
Hare NJ, Ma CS, Alvaro F, Nichols KE, Tangye SG (2006) Missense mutations in SH2D1A identified in patients with X-linked lymphoproliferative disease differentially affect the expression and function of SAP. Int Immunol 18:1055–1065
Sayos J, Wu C, Morra M, Wang N, Zhang X, Allen D, van Schaik S, Notarangelo L, Geha R, Roncarolo MG, Oettgen H, De Vries JE, Aversa G, Terhorst C (1998) The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature 395:462–469
Hwang PM, Li C, Morra M, Lillywhite J, Muhandiram DR, Gertler F, Terhorst C, Kay LE, Pawson T, Forman-Kay JD, Li SC (2002) A “three-pronged” binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome. EMBO J 21:314–323
Finerty PJ, Muhandiram R, Forman-Kay JD (2002) Side-chain dynamics of the SAP SH2 domain correlate with a binding hot spot and a region with conformational plasticity. J Mol Biol 322:605–620
Eissmann P, Beauchamp L, Wooters J, Tilton JC, Long EO, Watzl C (2005) Molecular basis for positive and negative signaling by the natural killer cell receptor 2B4 (CD244). Blood 105:4722–4729
Morra M, Lu J, Poy F, Martin M, Sayos J, Calpe S, Gullo C, Howie D, Rietdijk S, Thompson A, Coyle AJ, Denny C, Yaffe MB, Engel P, Eck MJ, Terhorst C (2001) Structural basis for the interaction of the free SH2 domain EAT-2 with SLAM receptors in hematopoietic cells. EMBO J 20:5840–5852
Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, Howie D, Sumegi J, Terhorst C, Eck MJ (2003) SAP couples Fyn to SLAM immune receptors. Nat Cell Biol 5:155–160
Seemayer TA, Gross TG, Egeler RM, Pirruccello SJ, Davis JR, Kelly CM, Okano M, Lanyi A, Sumegi J (1995) X-linked lymphoproliferative disease: twenty-five years after the discovery. Pediatr Res 38:471–478
Latour S, Roncagalli R, Chen R, Bakinowski M, Shi X, Schwartzberg PL, Davidson D, Veillette A (2003) Binding of SAP SH2 domain to FynT SH3 domain reveals a novel mechanism of receptor signalling in immune regulation. Nat Cell Biol 5:149–154
Simarro M, Lanyi A, Howie D, Poy F, Bruggeman J, Choi M, Sumegi J, Eck MJ, Terhorst C (2004) SAP increases FynT kinase activity and is required for phosphorylation of SLAM and Ly9. Int Immunol 16:727–736
Chen R, Latour S, Shi X, Veillette A (2006) Association between SAP and FynT: inducible SH3 domain-mediated interaction controlled by engagement of the SLAM receptor. Mol Cell Biol 26:5559–5568
Li W, Sofi MH, Rietdijk S, Wang N, Terhorst C, Chang CH (2007) The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity 27:763–774
Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, Bendelac A (2007) Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity 27:751–762
Cannons JL, Yu LJ, Hill B, Mijares LA, Dombroski D, Nichols KE, Antonellis A, Koretzky GA, Gardner K, Schwartzberg PL (2004) SAP regulates T(H)2 differentiation and PKC-theta-mediated activation of NF-kappaB1. Immunity 21:693–706
Borowski C, Bendelac A (2005) Signaling for NKT cell development: the SAP–FynT connection. J Exp Med 201:833–836
Gu C, Tangye SG, Sun X, Luo Y, Lin Z, Wu J (2006) The X-linked lymphoproliferative disease gene product SAP associates with PAK-interacting exchange factor and participates in T cell activation. Proc Natl Acad Sci 103:14447–14452
Czar MJ, Kersh EN, Mijares LA, Lanier G, Lewis J, Yap G, Chen A, Sher A, Duckett CS, Ahmed R, Schwartzberg PL (2001) Altered lymphocyte responses and cytokine production in mice deficient in the X-linked lymphoproliferative disease gene SH2D1A/DSHP/SAP. Proc Natl Acad Sci USA 98:7449–7454
Crotty S, McCausland MM, Aubert RD, Wherry EJ, Ahmed R (2006) Hypogammaglobulinemia and exacerbated CD8 T-cell-mediated immunopathology in SAP-deficient mice with chronic LCMV infection mimics human XLP disease. Blood 108:3085–3093
Chen G, Tai AK, Lin M, Chang F, Terhorst C, Huber BT (2005) Signaling lymphocyte activation molecule-associated protein is a negative regulator of the CD8 T cell response in mice. J Immunol 175:2212–2218
Yin L, Al Alem U, Liang J, Tong WM, Li C, Badiali M, Medard JJ, Sumegi J, Wang ZQ, Romeo G (2003) Mice deficient in the X-linked lymphoproliferative disease gene sap exhibit increased susceptibility to murine gammaherpesvirus-68 and hypo-gammaglobulinemia. J Med Virol 71:446–455
Graham DB, Bell MP, McCausland MM, Huntoon CJ, van Deursen J, Faubion WA, Crotty S, McKean DJ (2006) Ly9 (CD229)-deficient mice exhibit T cell defects yet do not share several phenotypic characteristics associated with. J Immunol 176:291–300
Howie D, Laroux FS, Morra M, Satoskar AR, Rosas LE, Faubion WA, Julien A, Rietdijk S, Coyle AJ, Fraser C, Terhorst C (2005) Cutting edge: the SLAM family receptor Ly108 controls T cell and neutrophil functions. J Immunol 174:5931–5935
Wang N, Satoskar A, Faubion W, Howie D, Okamoto S, Feske S, Gullo C, Clarke K, Sosa MR, Sharpe AH, Terhorst C (2004) The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med 199:1255–1264
Chen G, Tai AK, Lin M, Chang F, Terhorst C, Huber BT (2007) Increased proliferation of CD8+ T cells in SAP-deficient mice is associated with impaired activation-induced cell death. Eur J Immunol 37:663–674
Nagy N, Matskova L, Kis LL, Hellman U, Klein G, Klein E (2009) The proapoptotic function of SAP provides a clue to the clinical picture of X-linked lymphoproliferative disease. Proc Natl Acad Sci USA 106:11966–11971
Snow AL, Marsh RA, Krummey SM, Roehrs P, Young LR, Zhang K, van Hoff J, Dhar D, Nichols KE, Filipovich AH, Su HC, Bleesing JJ, Lenardo MJ (2009) Restimulation-induced apoptosis of T cells is impaired in patients with X-linked lymphoproliferative disease caused by SAP deficiency. J Clin Invest 119:2976–2989
Crotty S, Kersh EN, Cannons J, Schwartzberg PL, Ahmed R (2003) SAP is required for generating long-term humoral immunity. Nature 421:282–287
Morra M, Barrington RA, Abadia-Molina AC, Okamoto S, Julien A, Gullo C, Kalsy A, Edwards MJ, Chen G, Spolski R, Leonard WJ, Huber BT, Borrow P, Biron CA, Satoskar AR, Carroll MC, Terhorst C (2005) Defective B cell responses in the absence of SH2D1A. Proc Natl Acad Sci USA 102:4819–4823
Cannons JL, Yu LJ, Jankovic D, Crotty S, Horai R, Kirby M, Anderson S, Cheever AW, Sher A, Schwartzberg PL (2006) SAP regulates T cell-mediated help for humoral immunity by a mechanism distinct from cytokine regulation. J Exp Med 203:1551–1565
Hron JD, Caplan L, Gerth AJ, Schwartzberg PL, Peng SL (2004) SH2D1A regulates T-dependent humoral autoimmunity. J Exp Med 200:261–266
Ma CS, Pittaluga S, Avery DT, Hare NJ, Maric I, Klion AD, Nichols KE, Tangye SG (2006) Selective generation of functional somatically mutated IgM+CD27+, but not Ig isotype-switched, memory B cells in X-linked lymphoproliferative disease. J Clin Invest 116:322–333
Kamperschroer C, Dibble JP, Meents DL, Schwartzberg PL, Swain SL (2006) SAP is required for Th cell function and for immunity to influenza. J Immunol 177:5317–5327
Ma CS, Hare NJ, Nichols KE, Dupre L, Andolfi G, Roncarolo MG, Adelstein S, Hodgkin PD, Tangye SG (2005) Impaired humoral immunity in X-linked lymphoproliferative disease is associated with defective IL-10 production by CD4+ T cells. J Clin Invest 115:1049–1059
Malbran A, Belmonte L, Ruibal-Ares B, Bare P, Massud I, Parodi C, Felippo M, Hodinka R, Haines K, Nichols KE, de Bracco MM (2004) Loss of circulating CD27+ memory B cells and CCR4+ T cells occurring in association with elevated EBV loads in XLP patients surviving primary EBV infection. Blood 103:1625–1631
Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN (2008) SAP-controlled T–B cell interactions underlie germinal centre formation. Nature 455:764–769
Kamperschroer C, Roberts DM, Zhang Y, Weng NP, Swain SL (2008) SAP enables T cells to help B cells by a mechanism distinct from Th cell programming or CD40 ligand regulation. J Immunol 181:3994–4003
Schwartzberg PL, Mueller KL, Qi H, Cannons JL (2009) SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat Rev Immunol 9:39–46
McHeyzer-Williams LJ, Pelletier N, Mark L, Fazilleau N, McHeyzer-Williams MG (2009) Follicular helper T cells as cognate regulators of B cell immunity. Curr Opin Immunol 21:266–273
King C, Tangye SG, Mackay CR (2008) T follicular helper (TFH) cells in normal and dysregulated immune responses. Annu Rev Immunol 26:741–766
Fazilleau N, Mark L, McHeyzer-Williams LJ, McHeyzer-Williams MG (2009) Follicular helper T cells: lineage and location. Immunity 30:324–335
Eddahri F, Denanglaire S, Bureau F, Spolski R, Leonard WJ, Leo O, Andris F (2009) Interleukin-6/STAT3 signaling regulates the ability of naive T cells to acquire B-cell help capacities. Blood 113:2426–2433
Wu C, Nguyen KB, Pien GC, Wang N, Gullo C, Howie D, Sosa MR, Edwards MJ, Borrow P, Satoskar AR, Sharpe AH, Biron CA, Terhorst C (2001) SAP controls T cell responses to virus and terminal differentiation of TH2 cells. Nat Immunol 2:410–414
Davidson D, Shi X, Zhang S, Wang H, Nemer M, Ono N, Ohno S, Yanagi Y, Veillette A (2004) Genetic evidence linking SAP, the X-linked lymphoproliferative gene product, to Src-related kinase FynT in T(H)2 cytokine regulation. Immunity 21:707–717
McCausland MM, Yusuf I, Tran H, Ono N, Yanagi Y, Crotty S (2007) SAP regulation of follicular helper CD4 T cell development and humoral immunity is independent of SLAM and Fyn kinase. J Immunol 178:817–828
Kronenberg M, Kinjo Y (2009) Innate-like recognition of microbes by invariant natural killer T cells. Curr Opin Immunol 21:391–396
Bendelac A, Savage PB, Teyton L (2007) The biology of NKT cells. Annu Rev Immunol 25:297–336
Godfrey DI, McCluskey J, Rossjohn J (2005) CD1d antigen presentation: treats for NKT cells. Nat Immunol 6:754–756
Godfrey DI, Kronenberg M (2004) Going both ways: immune regulation via CD1d-dependent NKT cells. J Clin Invest 114:1379–1388
Pichavant M, Goya S, Meyer EH, Johnston RA, Kim HY, Matangkasombut P, Zhu M, Iwakura Y, Savage PB, Dekruyff RH, Shore SA, Umetsu DT (2008) Ozone exposure in a mouse model induces airway hyperreactivity that requires the presence of natural killer T cells and IL-17. J Exp Med 205:385–393
Chung B, Aoukaty A, Dutz J, Terhorst C, Tan R (2005) Cutting edge: signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J Immunol 174:3153–3157
Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, Stein PL (2005) Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med 11:340–345
Pasquier B, Yin L, Fondaneche MC, Relouzat F, Bloch-Queyrat C, Lambert N, Fischer A, Saint-Basile G, Latour S (2005) Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med 201:695–701
Eberl G, Lowin-Kropf B, MacDonald HR (1999) Cutting edge: NKT cell development is selectively impaired in Fyn-deficient mice. J Immunol 163:4091–4094
Gadue P, Morton N, Stein PL (1999) The Src family tyrosine kinase Fyn regulates natural killer T cell development. J Exp Med 190:1189–1196
Nunez-Cruz S, Yeo WC, Rothman J, Ojha P, Bassiri H, Juntilla M, Davidson D, Veillette A, Koretzky GA, Nichols KE (2008) Differential requirement for the SAP–Fyn interaction during NK T cell development and function. J Immunol 181:2311–2320
Gapin L, Matsuda JL, Surh CD, Kronenberg M (2001) NKT cells derive from double-positive thymocytes that are positively selected by CDId. Nature Immunol 2:971–978
Zhou D, Mattner J, Cantu C III, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, Teneberg S, Wang D, Proia RL, Levery SB, Savage PB, Teyton L, Bendelac A (2004) Lysosomal glycosphingolipid recognition by NKT cells. Science 306:1786–1789
Schmidt-Supprian M, Tian J, Grant EP, Pasparakis M, Maehr R, Ovaa H, Ploegh HL, Coyle AJ, Rajewsky K (2004) Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-kappaB activation. Proc Natl Acad Sci USA 101:4566–4571
Sivakumar V, Hammond KJ, Howells N, Pfeffer K, Weih F (2003) Differential requirement for Rel/nuclear factor [kappa]B family members in natural killer T cell development. J Exp Med 197:1613–1621
Diana J, Griseri T, Lagaye S, Beaudoin L, Autrusseau E, Gautron AS, Tomkiewicz C, Herbelin A, Barouki R, von Herrath M, Dalod M, Lehuen A (2009) NKT cell-plasmacytoid dendritic cell cooperation via OX40 controls viral infection in a tissue-specific manner. Immunity 30:289–299
Roberts TJ, Lin Y, Spence PM, Van Kaer L, Brutkiewicz RR (2004) CD1d1-dependent control of the magnitude of an acute antiviral immune response. J Immunol 172:3454–3461
Yuling H, Ruijing X, Li L, Xiang J, Rui Z, Yujuan W, Lijun Z, Chunxian D, Xinti T, Wei X, Lang C, Yanping J, Tao X, Mengjun W, Jie X, Youxin J, Jinquan T (2009) EBV-induced human CD8+ NKT cells suppress tumorigenesis by EBV-associated malignancies. Cancer Res 69:7935
Terabe M, Berzofsky JA (2008) The role of NKT cells in tumor immunity. Adv Cancer Res 101:277–348
Terabe M, Berzofsky JA (2007) NKT cells in immunoregulation of tumor immunity: a new immunoregulatory axis. Trends Immunol 28:491–496
Galli G, Pittoni P, Tonti E, Malzone C, Uematsu Y, Tortoli M, Maione D, Volpini G, Finco O, Nuti S, Tavarini S, Dellabona P, Rappuoli R, Casorati G, Abrignani S (2007) Invariant NKT cells sustain specific B cell responses and memory. Proc Natl Acad Sci USA 104:3984–3989
Lang GA, Devera TS, Lang ML (2008) Requirement for CD1d expression by B cells to stimulate NKT cell-enhanced antibody production. Blood 111:2158–2162
Tonti E, Galli G, Malzone C, Abrignani S, Casorati G, Dellabona P (2009) NKT-cell help to B lymphocytes can occur independently of cognate interaction. Blood 113:370–376
Cen O, Ueda A, Guzman L, Jain J, Bassiri H, Nichols KE, Stein PL (2009) The adaptor molecule signaling lymphocytic activation molecule-associated protein (SAP) regulates IFN-gamma and IL-4 production in V alpha 14 transgenic NKT cells via effects on GATA-3 and T-bet expression. J Immunol 182:1370–1378
Bassiri H, Janice Yeo WC, Rothman J, Koretzky GA, Nichols KE (2008) X-linked lymphoproliferative disease (XLP): a model of impaired anti-viral, anti-tumor and humoral immune responses. Immunol Res 42:145–159
Berg LJ (2007) Signalling through TEC kinases regulates conventional versus innate CD8+ T-cell development. Nat Rev Immunol 7:479–485
Prince AL, Yin CC, Enos ME, Felices M, Berg LJ (2009) The Tec kinases Itk and Rlk regulate conventional versus innate T-cell development. Immunol Rev 228:115–131
Lucas JA, Atherly LO, Berg LJ (2002) The absence of Itk inhibits positive selection without changing lineage commitment. J Immunol 168:6142–6151
Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ (2006) The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity 25:79–91
Readinger JA, Mueller KL, Venegas AM, Horai R, Schwartzberg PL (2009) Tec kinases regulate T-lymphocyte development and function: new insights into the roles of Itk and Rlk/Txk. Immunol Rev 228:93–114
Dubois S, Waldmann TA, Muller JR (2006) ITK and IL-15 support two distinct subsets of CD8+ T cells. Proc Natl Acad Sci 103:12075–12080
Broussard C, Fleischecker C, Horai R, Chetana M, Venegas AM, Sharp LL, Hedrick SM, Fowlkes BJ, Schwartzberg PL (2006) Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity 25:93–104
Horai R, Mueller KL, Handon RA, Cannons JL, Anderson SM, Kirby MR, Schwartzberg PL (2007) Requirements for selection of conventional and innate T lymphocyte lineages. Immunity 27:775–785
Li W, Kim MG, Gourley TS, McCarthy BP, Sant’Angelo DB, Chang CH (2005) An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity 23:375–386
Choi EY, Jung KC, Park HJ, Chung DH, Song JS, Yang SD, Simpson E, Park SH (2005) Thymocyte–thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity 23:387–396
Hu J, August A (2008) Naive and innate memory phenotype CD4+ T cells have different requirements for active Itk for their development. J Immunol 180:6544–6552
Yan Q, Malashkevich VN, Fedorov A, Fedorov E, Cao E, Lary JW, Cole JL, Nathenson SG, Almo SC (2007) Structure of CD84 provides insight into SLAM family function. Proc Natl Acad Sci USA 104:10583–10588
Concannon P, Rich SS, Nepom GT (2009) Genetics of type 1A diabetes. N Engl J Med 360:1646–1654
Coenen MJ, Gregersen PK (2009) Rheumatoid arthritis: a view of the current genetic landscape. Genes Immun 10:101–111
Zuvich RL, McCauley JL, Pericak-Vance MA, Haines JL (2009) Genetics and pathogenesis of multiple sclerosis. Semin Immunol 21:328–333
Krishnan S, Chowdhury B, Tsokos GC (2006) Autoimmunity in systemic lupus erythematosus: integrating genes and biology. Semin Immunol 18:230–243
Moser KL, Gray-McGuire C, Kelly J, Asundi N, Yu H, Bruner GR, Mange M, Hogue R, Neas BR, Harley JB (1999) Confirmation of genetic linkage between human systemic lupus erythematosus and chromosome 1q41. Arthritis Rheum 42:1902–1907
Shai R, Quismorio FP Jr, Li L, Kwon OJ, Morrison J, Wallace DJ, Neuwelt CM, Brautbar C, Gauderman WJ, Jacob CO (1999) Genome-wide screen for systemic lupus erythematosus susceptibility genes in multiplex families. Hum Mol Genet 8:639–644
Tsao BP, Cantor RM, Grossman JM, Kim SK, Strong N, Lau CS, Chen CJ, Shen N, Ginzler EM, Goldstein R, Kalunian KC, Arnett FC, Wallace DJ, Hahn BH (2002) Linkage and interaction of loci on 1q23 and 16q12 may contribute to susceptibility to systemic lupus erythematosus. Arthritis Rheum 46:2928–2936
Tsao BP, Cantor RM, Kalunian KC, Chen CJ, Badsha H, Singh R, Wallace DJ, Kitridou RC, Chen SL, Shen N, Song YW, Isenberg DA, Yu CL, Hahn BH, Rotter JI (1997) Evidence for linkage of a candidate chromosome 1 region to human systemic lupus erythematosus. J Clin Invest 99:725–731
Johanneson B, Lima G, von Salome J, Alarcon-Segovia D, Alarcon-Riquelme ME (2002) A major susceptibility locus for systemic lupus erythematosus maps to chromosome 1q31. Am J Hum Genet 71:1060–1071
Gray-McGuire C, Moser KL, Gaffney PM, Kelly J, Yu H, Olson JM, Jedrey CM, Jacobs KB, Kimberly RP, Neas BR, Rich SS, Behrens TW, Harley JB (2000) Genome scan of human systemic lupus erythematosus by regression modeling: evidence of linkage and epistasis at 4p16–15.2. Am J Hum Genet 67:1460–1469
Wakeland EK, Liu K, Graham RR, Behrens TW (2001) Delineating the genetic basis of systemic lupus erythematosus. Immunity 15:397–408
Vyse TJ, Richardson AM, Walsh E, Farwell L, Daly MJ, Terhorst C, Rioux JD (2005) Mapping autoimmune disease genes in humans: lessons from IBD and SLE. Novartis Found Symp 267:94–107
Graham DSC, Vyse TJ, Fortin PR, Montpetit A, Cai Y, Lim S, McKenzie T, Farwell L, Rhodes B, Chad L, Hudson TJ, Sharpe A, Terhorst C, Greenwood CMT, Wither J, Rioux JD (2008) Association of LY9 in UK and Canadian SLE families. Genes Immun 9:93–102
Sayos J, Martin M, Chen A, Simarro M, Howie D, Morra M, Engel P, Terhorst C (2001) Cell surface receptors Ly-9 and CD84 recruit the X-linked lymphoproliferative disease gene product SAP. Blood 97:3867–3874
Rozzo S, Vyse T, Drake C, Kotzin B (1996) Effect of genetic background on the contribution of New Zealand Black loci to autoimmune lupus nephritis. Proc Natl Acad Sci 93:15164–15168
Kono DH, Burlingame RW, Owens DG, Kuramochi A, Balderas RS, Balomenos D, Theofilopoulos AN (1994) Lupus susceptibility loci in New Zealand mice. Proc Natl Acad Sci 91:10168–10172
Hogarth MB, Slingsby JH, Allen PJ, Thompson EM, Chandler P, Davies KA, Simpson E, Morley BJ, Walport MJ (1998) Multiple lupus susceptibility loci map to chromosome 1 in BXSB mice. J Immunol 161:2753–2761
Morel L, Blenman KR, Croker BP, Wakeland EK (2001) The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc Natl Acad Sci 98:1787–1792
Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR Jr, Morel L, Wakeland EK (2004) Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity 21:769–780
Chen YF, Perry D, Boackle SA, Sobel ES, Molina H, Croker BP, Morel L (2005) Several genes contribute to the production of autoreactive B and T cells in the murine lupus susceptibility locus Sle1c. J Immunol 175:1080–1089
Chen YF, Cuda C, Morel L (2005) Genetic determination of T cell help in loss of tolerance to nuclear antigens. J Immunol 174:7692–7702
Croker BP, Gilkeson G, Morel L (2003) Genetic interactions between susceptibility loci reveal epistatic pathogenic networks in murine lupus. Genes Immun 4:575–585
Bygrave AE, Rose KL, Cortes-Hernandez J, Warren J, Rigby RJ, Cook HT, Walport MJ, Vyse TJ, Botto M (2004) Spontaneous autoimmunity in 129 and C57BL/6 mice—implications for autoimmunity described in gene-targeted mice. PLoS Biol 2:E243
Carlucci F, Cortes-Hernandez J, Fossati-Jimack L, Bygrave AE, Walport MJ, Vyse TJ, Cook HT, Botto M (2007) Genetic dissection of spontaneous autoimmunity driven by 129-derived chromosome 1 Loci when expressed on C57BL/6 mice. J Immunol 178:2352–2360
Chen YF, Morel L (2005) Genetics of T cell defects in lupus. Cell Mol Immunol 2:403–409
Kaminski DA, Stavnezer J (2007) Antibody class switching differs among SJL, C57BL/6 and 129 mice. Int Immunol 19:545–556
Corcoran LM, Metcalf D (1999) IL-5 and Rp105 signaling defects in B cells from commonly used 129 mouse substrains. J Immunol 163:5836–5842
McVicar DW, Winkler-Pickett R, Taylor LS, Makrigiannis A, Bennett M, Anderson SK, Ortaldo JR (2002) Aberrant DAP12 signaling in the 129 strain of mice: implications for the analysis of gene-targeted mice. J Immunol 169:1721–1728
O’Neill SM, Brady MT, Callanan JJ, Mulcahy G, Joyce P, Mills KH, Dalton JP (2000) Fasciola hepatica infection downregulates Th1 responses in mice. Parasite Immunol 22:147–155
Mo XY, Sangster M, Sarawar S, Coleclough C, Doherty PC (1995) Differential antigen burden modulates the gamma interferon but not the immunoglobulin response in mice that vary in susceptibility to Sendai virus pneumonia. J Virol 69:5592–5598
Fritz RB, Zhao ML (1996) Active and passive experimental autoimmune encephalomyelitis in strain 129/J (H-2b) mice. J Neurosci Res 45:471–474
Mittleman BB, Morse HC III, Payne SM, Shearer GM, Mozes E (1996) Amelioration of experimental systemic lupus erythematosus (SLE) by retrovirus infection. J Clin Immunol 16:230–236
Morel L, Blenman KR, Croker BP, Wakeland EK (2001) The major murine systemic lupus erythematosus susceptibility locus, Sle1, is a cluster of functionally related genes. Proc Natl Acad Sci USA 98:1787–1792
Xie C, Patel R, Wu T, Zhu J, Henry T, Bhaskarabhatla M, Samudrala R, Tus K, Gong Y, Zhou H, Wakeland EK, Zhou XJ, Mohan C (2007) PI3K/AKT/mTOR hypersignaling in autoimmune lymphoproliferative disease engendered by the epistatic interplay of Sle1b and FASlpr. Int Immunol 19:509–522
Kumar KR, Li L, Yan M, Bhaskarabhatla M, Mobley AB, Nguyen C, Mooney JM, Schatzle JD, Wakeland EK, Mohan C (2006) Regulation of B cell tolerance by the lupus susceptibility gene Ly108. Science 312:1665–1669
Jennings P, Chan A, Schwartzberg P, Wakeland EK, Yuan D (2008) Antigen-specific responses and ANA production in B6.Sle1b mice: a role for SAP. J Autoimmun 31:345–353
Komori H, Furukawa H, Mori S, Ito MR, Terada M, Zhang MC, Ishii N, Sakuma N, Nose M, Ono M (2006) A signal adaptor SLAM-associated protein regulates spontaneous autoimmunity and Fas-dependent lymphoproliferation in MRL-Faslpr lupus mice. J Immunol 176:395–400
Linterman MA, Rigby RJ, Wong RK, Yu D, Brink R, Cannons JL, Schwartzberg PL, Cook MC, Walters GD, Vinuesa CG (2009) Follicular helper T cells are required for systemic autoimmunity. J Exp Med 206:561–576
Acknowledgements
We are grateful for support from the NIH (AI-15066, AI-076210, AI-065687 to CT) and the CCFA (to XR).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Detre, C., Keszei, M., Romero, X. et al. SLAM family receptors and the SLAM-associated protein (SAP) modulate T cell functions. Semin Immunopathol 32, 157–171 (2010). https://doi.org/10.1007/s00281-009-0193-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00281-009-0193-0