Abstract
Purpose
To investigate the mechanistic basis of the anti-tumor effect of the compound ITB-301.
Methods
Chemical modifications of genistein have been introduced to improve its solubility and efficacy. The anti-tumor effects were tested in ovarian cancer cells using proliferation assays, cell cycle analysis, immunofluorescence, and microscopy.
Results
In this work, we show that a unique glycoside of genistein, ITB-301, inhibits the proliferation of SKOv3 ovarian cancer cells. We found that the 50% growth inhibitory concentration of ITB-301 in SKOv3 cells was 0.5 μM. Similar results were obtained in breast cancer, ovarian cancer, and acute myelogenous leukemia cell lines. ITB-301 induced significant time- and dose-dependent microtubule depolymerization. This depolymerization resulted in mitotic arrest and inhibited proliferation in all ovarian cancer cell lines examined including SKOv3, ES2, HeyA8, and HeyA8-MDR cells. The cytotoxic effect of ITB-301 was dependent on its induction of mitotic arrest as siRNA-mediated depletion of BUBR1 significantly reduced the cytotoxic effects of ITB-301, even at a concentration of 10 μM. Importantly, efflux-mediated drug resistance did not alter the cytotoxic effect of ITB-301 in two independent cancer cell models of drug resistance.
Conclusion
These results identify ITB-301 as a novel anti-tubulin agent that could be used in cancers that are multidrug resistant. We propose a structural model for the binding of ITB-301 to α- and β-tubulin dimers on the basis of molecular docking simulations. This model provides a rationale for future work aimed at designing of more potent analogs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Drugs that effectively interfere with tubulin dynamics prevent mitosis in cancer cells, leading to cell cycle arrest and, eventually, apoptosis. These drugs are generally classified into two major groups: (1) microtubule stabilizers, such as taxanes and epothilones that induce a net polymerizing effect and (2) microtubule depolymerizers, such as the vinca alkaloids colchicine and nocodazole that induce a net depolymerizing effect [1]. Both types of tubulin inhibitors have been highly effective in the treatment of a variety of tumors [2, 3]. However, cancer cells can acquire resistance to these drugs through multiple mechanisms, including alterations in microtubule dynamics [4], overexpression of multidrug-resistance genes [5], delay of the G2/M transition [6], defects in mitotic checkpoints [7], and alterations in apoptotic pathways [8]. Thus, the development of novel anti-tubulin drugs that can escape these acquired resistance mechanisms is clinically important. ITB-301 (also called G21) is a novel lipophilic glycoside derivative of genistein that has been shown to possess potent anti-proliferative activity in cancer cells through undetermined mechanisms [9]. In this work, we show that ITB-301 [10] induces severe microtubule depolymerization and inhibits cancer-cell proliferation. Importantly, efflux-mediated drug resistance did not affect the efficacy of ITB-301, indicating that ITB-301 is potentially useful in treating multidrug-resistant tumors.
Materials and methods
Materials
Compound ITB-301 (Fig. 1a) was a gift from IntertechBio Corporation and was synthesized using a previously reported method [9].
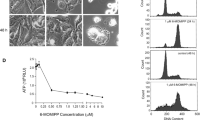
The lipophilic disaccharide moiety of genistein significantly increases its cytotoxicity. a The structures of genistein and its glycoside derivative, ITB-301. b SKOv3 and ES2 ovarian cancer cells were cultured in 96-well plates (5,000 cells per well) and allowed to adhere overnight. The cells were then treated with the drugs indicated at nine different concentrations for 48 h before being fixed and stained. The percent proliferation compared with untreated cells after 48 h of drug treatment was calculated. Shown is the mean percent proliferation (±SEM) at each drug concentration calculated from at least six replicates per concentration
Cell lines
All cell lines, excluding the SKOv3-TR cell line, were obtained from the American Tissue Type Culture Collection. The SKOv3-TR cell line was obtained from Cell Services (Cancer Research UK London Research Institute). Cells were maintained in RPMI-140 medium supplemented with 10% fetal bovine serum and incubated at 37°C and 5% CO2. The SKOv3-TR cells were maintained in RPMI supplemented with 0.3 μM paclitaxel.
Proliferation and apoptosis assays
To measure cell proliferation by crystal violet staining, cancer cells were plated at 5,000 cells per well in 96-well plates and allowed to adhere overnight. Cells were treated with ITB-301, paclitaxel (Sigma–Aldrich), or nocodazole (Sigma–Aldrich) at the concentrations indicated for 48 h. Verapamil (Sigma–Aldrich) was used at 10 μM in some experiments. For the estimation of the number of cells being treated at the time of drug application (time 0; T0), replicate plates were used in which cells were fixed rather than drug-treated. The cells were fixed using 1% glutaraldehyde and stained using 0.5% crystal violet in 20% methanol for 1 h before being thoroughly washed with water. The dye was solubilized using 100 μl of Sorenson’s buffer (0.9% sodium citrate, 0.02 N HCl, and 45% ethanol), and light absorbance was measured at 540 nm. To quantify the viability of HL60 cells, the Cell Titer Blue Assay (Promega) was used according to the manufacturer’s instructions. In brief, an equal volume of reagent was added to that of culture medium in each well and incubated for 3 h at 37°C and 5% CO2 followed by measurement of fluorescence signal from each well using a microtiter plate reader.
To detect apoptosis, cells (1,500 cells per well) were incubated in black 384-well plates (Corning), and caspase 3/7 activity was estimated using the Caspase-Glo 3/7 Assay Reagent (Promega) following the manufacturer’s instructions. In brief, an equal volume of reagent was added to that of culture medium in each well and incubated for 1 h at room temperature followed by measurement of the luminescence signal from each well using a microtiter plate reader.
Immunofluorescence and automated microscopy
Subconfluent cells growing on cover slips were fixed using 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 3–5 min. The cells were washed to remove the PFA and were incubated in absolute ethanol at −20°C for at least 1 h before being washed with tris-buffered saline, 0.04% SDS (Sigma), and 0.2% Triton X-100 (Sigma–Aldrich). Blocking was performed using 1.5% bovine serum albumin in tris-buffered saline. Mouse anti-α-tubulin antibody (Sigma–Aldrich) was incubated at a 1:100 dilution in blocking solution for 1 h at room temperature. The primary antibody was detected using Alexa-488- or Alexa-594-conjugated anti-mouse antibodies (Invitrogen). Nuclear DNA was stained using Hoechst 33258 dye (Invitrogen). Images were observed using an Olympus FV1000 microscope.
For automated microscopy, the same staining steps were performed in black 384-well plates (Corning). Eight images were collected per well using the In Cell Analyzer 1000 (GE Healthcare) with a Nikon 20× PlanFluo lens (Nikon), numerical aperture of 0.45, refractive index of 1 and a focal length of 10,000 pixel width and height; 0.323 and a linear type of 7 μm. A binning value of 1 × 1 and a gain value of 2 were used to generate the images (1,392 × 1,040). The images were analyzed with an In Cell Investigator software (GE Healthcare) using a multi-target analysis protocol. For detecting fluorescence signals, the excitation wavelength of 360 nm and emission of 535 nM were used for Hoechst stain with an exposure time of 200 ms and an adjustable offset value, typically at −9.2 μm, and 480/535 nm for Alexa-488 with a typical exposure time of 600 ms and an offset of −0.8 μm. A multi-target analysis protocol was used for segmentation. Microtubule pixel intensity was extracted using a top-hat segmentation method specifying a minimum cell area of 100 μm2 and sensitivity for detecting pixel clusters of 74%. In all high-content experiments, microtubule segmentation was reviewed in at least 30 randomly selected fields to check for appropriate segmentation.
Tubulin in vitro polymerization assay
Measuring in vitro polymerization of tubulin was performed by monitoring turbidity changes in tubulin spectrophotometrically at A340 over 60 min using the tubulin polymerization assay kit (Cytoskeleton) according to manufacturer’s instructions. In brief, 100 μl volume of 3 mg/ml tubulin in 80 mM PIPES pH 7.0, 0.5 mM EGTA, 2 mM MgCl2, 1 mM GTP, and 10% glycerol was treated with either DMSO (0.5%), ITB-301 10 μM, paclitaxel 10 μM or Nocodazole 10 μM in 96-well plates. Polymerization was started by incubation at 37°C and followed by absorption readings at 340 nm using the Fusion universal microplate analyzer (Perkin Elmer). All treatments were performed in triplicate. To estimate the predicted maximum tubulin polymer mass, a non-linear curve was fitted to the time-series data using Michaelis–Menten model (Graphpad prism 5 software, GraphPad Software, Inc). Using this method, a goodness of fit R 2 of 0.88 was obtained for fitting the data from DMSO-treated and ITB-301-treated tubulin. The alternative hypothesis that the Vmax was different for each data set was tested at alpha level of 0.05.
Statistical analysis
The 50% growth inhibitory concentration (GI 50) was estimated using the following formula: 100 × (T − T0)/(C − T0) = 50, where T is the optical density (OD) value after drug treatment, T0 is the OD value at time 0, and C is the OD value for the control treatment [11]. Time 0 was defined as the day the drug was administered. Following normalization of the OD values for each drug concentration using the formula described above, the data were fitted to a non-linear curve using Graphpad Prism 5 software (GraphPad Software, Inc.) to determine the drug concentration at which cell proliferation was inhibited by 50%.
Statistical analysis for automated microscopy was conducted using the R environment [12], and graphs were plotted using the Graphpad Prism software.
Flow cytometry
Cells were treated with the drugs indicated for 24 h and then trypsinized and fixed with 4% paraformaldehyde for 3 min followed by 70% ethanol at −20°C for at least 1 h. Cells were stained using propidium iodide (Sigma–Aldrich), processed using an LSR II flow cytometer (BD Bioscience), and analyzed using the FACSDiva software (BD Bioscience) to obtain a cell cycle profile. To estimate the mitotic index, the cells were fixed as described above for the cell cycle analysis and incubated with an anti-phospho-histone H3 antibody (Cell Signaling Technologies). The primary antibody was detected using an Alexa-488-tagged secondary antibody.
BUBR1 knockdown
Transient knockdown of BUBR1 was performed by transfecting SKOv3 cells with two different siRNA duplexes targeting BUBR1 or siControl non-targeting siRNA 5 (siGenome library, Dharmacon) using the Dharmafect 4 reagent (Dharmacon) according to the manufacturer’s instructions and as previously described [4]. In summary, 0.2 μl of 20 μM siRNA was mixed with 0.2 μl of Dharmafect 4 and 20 μl of Optimem medium (Invitrogen) for 20 min in a well of 384-well plate and overlaid with 80 μl of cell suspension (1,750 cells) in antibiotic-free cell line-specific media. This gave a final siRNA concentration of 40 nM per well. The sequences for the siRNAs used were as follows; BUBR1 siRNA 1, 5′-GGAAGAAGAUCUAGAUGUA-3′ and BUBR1 siRNA 2, 5′-CAAGAUGGCUGUAUUGUUU-3′.
Docking of ITB-301 to microtubules
Since microtubules are assembled from α- and β-tubulin dimers, in our studies, the structure of a model dimer was taken from the crystallographic data of α- and β-tubulin stabilized by paclitaxel [13], PDB code—1JFF. A model tubulin dimer complexed with colchicine was built using the crystallographic structure of β-tubulin complexed with colchicine [14], PDB code—ISA0, by replacing β-tubulin complexed with paclitaxel. The locations of colchicine and paclitaxel in their binding pockets are hereafter referred to as “the colchicine binding site” and “the paclitaxel binding site,” respectively. The MOE molecular modeling library [15] was used in the modeling studies. The reference structures were assigned hydrogen atoms using routine procedures implemented in MOE. Both structures were optimized by the energy minimization. Next, colchicine and paclitaxel were removed. Genistein was docked into the colchicine binding site, and ITB-301 was docked into the colchicine and paclitaxel binding sites using MOE docking technologies. Initial configurations were created manually using a 3D visualization system and by careful analysis of possible binding modes. In particular, selected positions of ITB-301 with its genistein fragment inside and outside the colchicine and paclitaxel binding sites were probed. The resulting possible binding modes were scored and relaxed by the energy minimization. Because ITB-301 is relatively large and its locations in the binding sites can be affected by interactions with the environment, the structural stability of the 16 most probable ITB-301-tubulin complexes in a water solution was tested by carrying out 1-ns molecular dynamics simulations, which were preceded by careful thermalization and equilibration of the systems. The above-mentioned simulations were carried out for the two model systems at T = 310 K, atmospheric pressure, and an ionic strength of 0.05 M in a rectangular cuboid. The first system consisted of α- and β-tubulin complexed with ITB-301 in the colchicine binding site and surrounded by 32,255 water molecules. The second system consisted of an α-β-α-tubulin trimer complexed with ITB-301 in the paclitaxel binding site and surrounded by 23,844 water molecules. This allowed for the drug to be located approximately in the center of the simulated system. The NAMD library [16, 17] capable of carrying out simulations for large molecular systems with scalable parallel MD algorithms was used. The CHARMM27 force field [18] and a TIP3P water model [19] were applied.
Results
ITB-301 inhibits cell proliferation and induces cell cycle arrest in ovarian cancer cells
In contrast to genistein, ITB-301 significantly inhibited the proliferation of SKOv3 ovarian cancer cells over the range of concentrations tested (0–10 μM, Fig. 1b). The GI50 of ITB-301 in SKOv3 cells was 0.52 μM (Fig. 2). Similar results were obtained for the ovarian cancer cell line ES2 and the breast cancer cell line HCC194 (Fig. 2). To investigate the cause of the cell-proliferation arrest following ITB-301 administration, we examined the cell cycle progression of SKOv3 cells, both before and after ITB-301 treatment, using flow cytometry. We found that 93% of the cells were arrested in the G2/M phase of the cell cycle 24 h after treatment, and 63% of these cells were arrested in mitosis, as evidenced by positive anti-phospho-histone H3 staining, which is a known mitosis marker (Fig. 3). The arrest of cells in mitosis by ITB-301 treatment is similar to the effect obtained when cells are treated with the anti-tubulin drugs paclitaxel and nocodazole (Fig. 3). Similar results were obtained in ES2, paclitaxel-sensitive HeyA8 and HeyA8 paclitaxel-resistant ovarian cancer cell lines (Supplementary Fig. 1).
ITB-301 induces significant cytotoxicity in ovarian (ES2) and breast cancer (HCC194) cell lines. Cancer cells were cultured in 96-well plates (5,000 cells per well) and allowed to adhere overnight. Cells were then treated with the indicated drugs at nine different concentrations for 48 h before being fixed and stained. The percent proliferation compared with untreated cells after 48 h of drug treatment was calculated. Shown is the mean percent proliferation (±SEM) calculated from at least six replicates per concentration from two independent experiments
ITB-301 induces mitotic arrest in SKOv3 cells. SKOv3 cells were treated with different drugs for 24 h before the cells were fixed and stained for cell cycle analysis using propidium iodide staining or the determination of the mitotic index using anti-phospho-histone H3 staining tagged with Alexa-488. Shown are typical results from two independent experiments
ITB-301 induces microtubule depolymerization
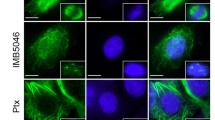
Because ITB-301 induced mitotic arrest similar to anti-tubulin agents, we hypothesized that ITB-301 treatment resulted in microtubule damage. To quantify the effect of ITB-301 on microtubules, we treated SKOv3 cells with 10 μM ITB-301 for 24 h and then examined the microtubule mass using automated microscopy and image analysis. The microtubule mass decreased significantly (P < 0.0001, t test) following ITB-301 treatment, and this was similar to the decrease in mass produced by treatment with 2 μM nocodazole. In contrast, treatment with 100 nM paclitaxel produced a significant increase in the microtubule mass (Fig. 4a, c–f). Treatment with ITB-301 induced a dose- and time-dependent decrease in the microtubule mass (Fig. 4b). Confocal microscopy was then used to qualitatively assess the effect of ITB-301 treatment on microtubules. Treatment of cells with ITB-301 at the GI50 concentration (0.5 μM) induced loss of microtubule structure within 1 h, in a manner similar to that achieved by treatment with nocodazole at the GI50 concentration (Fig. 4h and i). In contrast, paclitaxel induced the formation of characteristic microtubule bundles (Fig. 4g and j). Importantly, ITB-301 also induced microtubule depolymerization in ES2, paclitaxel-sensitive HeyA8, and HeyA8 paclitaxel-resistant ovarian cancer cell lines (Supplementary Fig. 1).
ITB-301 induces significant microtubule depolymerization. a and b SKOv3 cells were treated with drugs and then fixed and stained as described in the “Materials and methods”. Shown is the mean of the background (BG)-corrected intensity values (±SEM) for α-tubulin staining from at least 500 cells from triplicate wells. c–f Representative images from which the data in panel a were derived are shown. g–j Projected images of confocal microscopy Z-stacks of SKOv3 cells treated with the drugs indicated for 1 h before being fixed and stained using an α-tubulin antibody. Note the loss of microtubule architecture in panels h and i and the microtubule bundles in panel j (arrowhead). k 100 μl volume of 3 mg/ml tubulin was incubated with either 0.5% DMSO (control), 10 μM ITB-301, 10 μM paclitaxel, or 10 μM Nocodazole in 96-well plates in triplicates. Tubulin polymerization was started by incubation at 37°C and followed over 60 min by absorption readings at 340 nm. Scale bar = 5 μm
To test whether ITB-301 directly interacted with tubulin, in vitro polymerization assays were performed. Tubulin polymerization was determined in vitro by monitoring turbidity changes in tubulin spectrophotometrically at A340 over 60 min as an indirect measure of changes in microtubule mass. As expected, this showed that paclitaxel significantly enhanced microtubule polymerization (Fig. 4k). In contrast, nocodazole and ITB-301 decreased tubulin polymerization over time (Fig. 4k). To calculate the maximum predicted tubulin polymer mass (Vmax) over time, a non-linear curve was fitted to the time-series data (Michaelis–menten equation, Graphpad Prism). This analysis revealed that ITB-301 at 10 μM concentration significantly decreased the maximum predicted tubulin polymer mass over time by 12% compared with DMSO-treatment (predicted OD value of 0.22 and 0.25, respectively, P < 0.0001).
BUBR1 is required for ITB-301-induced cytotoxicity
Microtubule-targeting agents induce sustained activation of mitotic checkpoint regulators, such as BUBR1 and MAD2, resulting in apoptosis and cell death [20]. We hypothesized that ITB-301 induces microtubule depolymerization that results in mitotic checkpoint-dependent apoptosis and cell death. We first showed that ITB-301 induced apoptosis in SKOv3 cells in a dose-dependent manner (Fig. 5a). To test whether the ITB-301-induced apoptosis was dependent on intact mitotic checkpoints, we depleted BUBR1 in SKOV3 cells using two independent BUBR1-targeting siRNAs before treating cells with ITB-301. BUBR1 depletion diminished the ITB-301-induced apoptosis in SKOv3 cells at all three concentrations tested (P < 0.001, one-way ANOVA; Fig. 5b). Similar results were obtained using cell-proliferation assays (Fig. 5c). From these results, we concluded that the ITB-301-mediated cytotoxicity was dependent on active mitotic checkpoints.
BUBR1 is required for ITB-301-induced cytotoxicity. a SKOv3 cells were plated in a 384-well plate (2,000 cells per well) and permitted to adhere overnight. Cells were treated with ITB-301 at the concentrations shown for 48 h, and caspase 3/7 activity was determined. b SKOV3 cells were transfected using either two independent siRNAs targeting BUBR1 or non-targeting control siRNAs. Cells were treated with ITB-301 at the concentrations shown for 48 h, and caspase 3/7 activity was determined. The data shown in panels a and b represent the fold-change (±SD) in caspase 3/7 activity between the ITB-301-treated cells and the vehicle-treated controls. All values were measured in triplicate. c The cells were treated with siRNA as described in panel b, and the cell proliferation was estimated using crystal violet staining. Data shown are the mean percent cell proliferation at each drug concentration (±SD) normalized to vehicle-treated controls. All values were measured in triplicate
ITB-301 is not a target of drug-efflux mechanisms
Drug efflux has been shown to be a significant cause of clinical resistance to multiple drugs [21]. We asked whether this mode of drug resistance might also result in resistance to ITB-301. To test this possibility, we used an established model of efflux-mediated chemotherapy resistance: the parent acute myelogenous leukemia cell line HL60 and the daughter doxorubicin-resistant cell line HL60-dox. HL60-dox overexpresses the multidrug-resistant protein (MRP) [22, 23], whereas the parent line does not. As expected, the HL60-dox cells were highly resistant to paclitaxel. The minimum effective dose of paclitaxel that was required to significantly inhibit HL60-dox proliferation was at least 400-fold greater than that required to inhibit HL60 proliferation (from 12.5 nM for HL60 to 5 μM for HL60-dox; Fig. 6a). In contrast, there was no change in the minimum effective dose of ITB-301 (5 μM for both cell lines; Fig. 6b). This result indicated that the effectiveness of ITB-301 was not affected by the presence of the MRP gene product. Similar results were obtained in the ovarian cancer–sensitive HeyA8 cells and the derived paclitaxel-resistant HeyA8-MDR that is known to overexpress the MDR gene (Supplementary Fig 2). To further confirm this finding, we used a third model of resistance that is based on drug efflux: the parent paclitaxel-sensitive ovarian cancer cell line SKOv3 and the daughter paclitaxel-resistant cell line SKOV3-TR, which is known to overexpress MDR1 [4, 24]. The minimum effective dose of paclitaxel for the SKOV3-TR cells was at least 100-fold greater than that for the parent SKOV3 cells (1 μM and 10 nM, respectively; Fig. 6c). Co-treatment of cells with paclitaxel and verapamil, an MDR1 resistance-reversing agent, resensitized the SKOv3-TR cells to the 10-nM paclitaxel treatment. In contrast, the minimum effective dose (10 μM) of ITB-301 that significantly inhibited proliferation of SKOv3-TR cells (tenfold higher than the dose required for the parent SKOv3 cells) was not affected by the presence of verapamil (Fig. 6d). This result provided further confirmation that the efficacy of ITB-301 is not altered by drug-efflux resistance mechanisms.
Multidrug resistance does not significantly alter the cytotoxic effect of ITB-301. a and b The parental HL60 acute myelogenous leukemia cells and the HL60-derived MRP1-overexpressing cells (HL60-dox) were plated in a 384-well plate (2,000 cells per well) and treated with the drugs at the indicated concentrations for 48 h, and the cell viability was estimated as described in methods. Data shown are the mean percent cell viability at each drug concentration (±SD) normalized to vehicle-treated controls. All values were measured in triplicate. c and d The parental SKOv3 paclitaxel-sensitive cells and the SKOv3-derived paclitaxel-resistant cells (SKOv3-TR) were plated in 96-well plates and allowed to adhere overnight. The cells were treated with the indicated concentrations of either paclitaxel or ITB-301, with or without verapamil (10 μM) as indicated, for 72 h. The cells were fixed and stained using crystal violet. Data shown represent the mean percent cell growth at each drug concentration (±SD) normalized to vehicle-treated control cells. All values were measured in triplicate
Theoretical modeling of ITB-301 binding to tubulin supports a depolymerizing mechanism of action
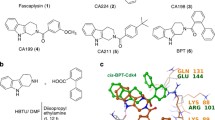
We next sought to compare the theoretical microtubule binding of ITB-301 to the binding of the known anti-tubulin drugs colchicine and paclitaxel. Packing ITB-301 in the tight paclitaxel binding site may result in local, steric hindrances and is, therefore, less likely to occur. A more plausible site that would accommodate the size and structure of ITB-301 may be between the α- and β-tubulin molecules. Colchicine, a known microtubule-depolymerizing agent binds at this site (Fig. 7a). Docking studies with MOE in implicit solvent have shown that (1) potential complexes of ITB-301 in the colchicine binding site were more probable than potential complexes of ITB-301 in the paclitaxel binding site and (2) positions of ITB-301 whereby its genistein fragment resides inside the binding pockets were typically more probable than those with the genistein fragment outside the binding pockets (Fig. 7b). These findings were refined by 1-ns molecular dynamics simulation runs in the explicit water solution starting from the 16 most probable cases selected from the docking studies. In each case, the mean interaction energies between ITB-301 and protein environments (including water molecules) in a sphere of 7 Å were computed. The residue-based cutoff criterion was applied. Complexes of ITB-301 in the colchicine binding site appeared to be more stable than those with ITB-301 in the paclitaxel binding site by more than 22 kcal/mol. The most stable complexes of ITB-301 with tubulin in the colchicine and the paclitaxel binding sites were selected. Their structures are depicted in Fig. 7c, d, respectively. The energy difference results mainly from unpreferable local interactions of ITB-301 in the paclitaxel binding site. These theoretical findings are in line with our experimental data, suggesting that intermolecular interactions of ITB-301 are more similar to that of colchicine than paclitaxel.
A structural model for binding of genistein and ITB-301 to tubulin dimers. a Structural models for binding of colchicine and paclitaxel to tubulin dimers are shown: α-tubulin (blue), β-tubulin (red), GTP (yellow), GDP (light blue), colchicine (green), and paclitaxel (white). b The three most probable positions of genistein in the colchicine binding site; the position shown in white has the largest overlap with the colchicine crystallographic location. GTP is colored yellow and the magnesium ion shown as an orange sphere. c The most stable position of ITB-301 (green) in the colchicine binding site. Note that the genistein fragment of ITB-301 penetrates the colchicine binding site, while the lipophilic disaccharide tail is located in the interface region between α- and β-tubulin. d Possible position of ITB-301 (green) in the paclitaxel binding site
Discussion
Genistein is one of the most intensively studied naturally occurring flavonoids and is commonly present in soy-derived products. It is generally accepted that genistein demonstrates considerable anticancer potential, with multiple mechanisms of action [25–27]. However, genistein’s low solubility in both aqueous and lipid environments limits its potential to be developed as a drug. Attempts to alter its bioavailability using derivatization with water-soluble molecules included the synthesis of sugar glycosides of genistein. Conjugation of genistein with a sugar derived from peracetylated lactose resulted in a compound, ITB-301, that was significantly more potent than other glycosides [9]. Surprisingly for a genistein derivative, our studies demonstrated for the first time that ITB-301 can induce significant dose- and time-dependent microtubule depolymerization [10]. A recent study provided further, although only qualitative, evidence of the effect of ITB-301 on microtubules [28]. In summary, our detailed studies of the mechanism of action of ITB-301 provided evidence that the cytotoxic effect of ITB-301 is almost exclusively the result of induction of microtubule depolymerization and subsequent mitotic arrest.
The precise mechanism of apoptosis induction following anti-tubulin drug treatment is unknown. However, there is strong evidence that intact mitotic checkpoints are crucial for the induction of mitotic arrest and apoptosis following anti-tubulin drug treatment [7, 20]. In this work, we found that depletion of BUBR1 nearly abolished the apoptosis induced by ITB-301. This finding confirms that, similar to anti-tubulin drugs, the cytotoxicity of ITB-301 requires intact mitotic checkpoints and is dependent on microtubule depolymerization.
How genistein and ITB-301 bind to tubulin and why ITB-301 induces depolymerization rather than stabilization remain to be identified. It is interesting to note that the genistein part of ITB-301 is capable of binding in the colchicine binding pocket at the interface between the α- and β-tubulin molecules. Binding of colchicine at this site requires a twist in the tubulin dimer conformation, and this prevents the necessary straight conformation that is needed for microtubule assembly [29]. While genistein alone seems to be capable of such binding, it is either incapable of or inefficient in inducing microtubule depolymerization. The lipophilic disaccharide tail of ITB-301 seems to be the key structural element allowing for effective microtubule depolymerization. Its location in the interface region between the α- and β-tubulin may either result in their separation or prevent the addition of new β-tubulin molecules at the microtubule end. This plausible model allows for a more rational design of novel derivatives of genistein with potent microtubule-depolymerizing properties and extends our knowledge of the mechanisms of tubulin drug binding and tubulin polymerization/depolymerization processes.
Acquired drug resistance via overexpression of transmembrane protein pumps that actively export drugs outside the cell is thought to be an important cause of clinical chemotherapy resistance [5, 23, 30]. Finding novel drugs with therapeutic effects similar to those of established chemotherapeutic agents but that are unaffected by drug-efflux pumps is highly desirable. Here, we showed that ITB-301 induced similar cytotoxic effects in both drug-sensitive leukemia cells and drug-resistant leukemia cells that overexpress MRP1 (HLA60-dox cells). In SKOv3-TR cells, we showed that the inhibition of MDR1 did not increase the cytotoxic effect of ITB-301, confirming that ITB-301 is not a substrate for MDR1-induced exocytosis. We speculate that the observed increase in ITB-301 resistance in SKOv3-TR may relate to other acquired resistance mechanisms. Our results indicate that ITB-301 could be used to treat cancers that have developed multidrug resistance. Interestingly, dimers of natural flavonoids, the same family to which genistein belongs, have recently been shown to inhibit MDR1-mediated drug efflux [31]. These dimers have been shown to reduce the IC50 of microtubule-targeting agents by up to 50-fold in models of drug resistance in vitro [31].
In summary, this work demonstrates that ITB-301 exhibits tubulin-depolymerizing characteristics against both drug-sensitive and multidrug-resistant cancer cell lines. Theoretical modeling of the mode of binding of ITB-301 to tubulin provides a rationale for the design of even more potent analogs.
References
Perez EA (2009) Microtubule inhibitors: differentiating tubulin-inhibiting agents based on mechanisms of action, clinical activity, and resistance. Mol Cancer Ther 8(8):2086–2095
McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, Clarke-Pearson DL, Davidson M (1996) Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N Engl J Med 334(1):1–6
Federico M, Luminari S, Iannitto E, Polimeno G, Marcheselli L, Montanini A, La Sala A, Merli F, Stelitano C, Pozzi S, Scalone R, Di Renzo N, Musto P, Baldini L, Cervetti G, Angrilli F, Mazza P, Brugiatelli M, Gobbi PG (2009) ABVD compared with BEACOPP compared with CEC for the initial treatment of patients with advanced Hodgkin’s lymphoma: results from the HD2000 gruppo italiano per lo studio dei linfomi trial. J Clin Oncol 27(5):805–811
Ahmed AA, Mills AD, Ibrahim AE, Temple J, Blenkiron C, Vias M, Massie CE, Iyer NG, McGeoch A, Crawford R, Nicke B, Downward J, Swanton C, Bell SD, Earl HM, Laskey RA, Caldas C, Brenton JD (2007) The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell 12(6):514–527
Kamazawa S, Kigawa J, Kanamori Y, Itamochi H, Sato S, Iba T, Terakawa N (2002) Multidrug resistance gene-1 is a useful predictor of paclitaxel-based chemotherapy for patients with ovarian cancer. Gynecol Oncol 86(2):171–176
Tan M, Jing T, Lan KH, Neal CL, Li P, Lee S, Fang D, Nagata Y, Liu J, Arlinghaus R, Hung MC, Yu D (2002) Phosphorylation on tyrosine-15 of p34(Cdc2) by ErbB2 inhibits p34(Cdc2) activation and is involved in resistance to taxol-induced apoptosis. Mol Cell 9(5):993–1004
Anand S, Penrhyn-Lowe S, Venkitaraman AR (2003) Aurora-a amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to taxol. Cancer Cell 3(1):51–62
Huang Y, Ray S, Reed JC, Ibrado AM, Tang C, Nawabi A, Bhalla K (1997) Estrogen increases intracellular p26Bcl-2 to p21Bax ratios and inhibits taxol-induced apoptosis of human breast cancer MCF-7 cells. Breast Cancer Res Treat 42(1):73–81
Polkowski K, Popiolkiewicz J, Krzeczynski P, Ramza J, Pucko W, Zegrocka-Stendel O, Boryski J, Skierski JS, Mazurek AP, Grynkiewicz G (2004) Cytostatic and cytotoxic activity of synthetic genistein glycosides against human cancer cell lines. Cancer Lett 203(1):59–69
Ahmed AA, Fokt I, Krzysko K, Lesyng B, Bast RC Jr, Priebe W (2009) Unsaturated glycoside of genistein induces severe microtubule depolymerization and mitotic arrest in ovarian cancer cells. In Proceedings of the AACR annual meeting, abstract number 5559
Monks A, Scudiero D, Skehan P, Shoemaker R, Paull K, Vistica D, Hose C, Langley J, Cronise P, Vaigro-Wolff A et al (1991) Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst 83(11):757–766
Ikada R, Gentleman R (1996) R: a language for data analysis and graphics. J Comp Graph Stat 5:299–314
Lowe J, Li H, Downing KH, Nogales E (2001) Refined structure of alpha beta-tubulin at 3.5 a resolution. J Mol Biol 313(5):1045–1057
Ravelli RB, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M (2004) Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 428(6979):198–202
Chemical Computing Group (2009) Molecular operating environment (moe). http://wwwchemcompcom/
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26(16):1781–1802
Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K (1999) NAMD2: greater scalability for parallel molecular dynamics. J Comput Physic 151:283–312
MacKerell AD Jr, Bashford D, Bellott M, Dunbrack RL Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher IW, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Sudo T, Nitta M, Saya H, Ueno NT (2004) Dependence of paclitaxel sensitivity on a functional spindle assembly checkpoint. Cancer Res 64(7):2502–2508
Gottesman MM, Fojo T, Bates SE (2002) Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer 2(1):48–58
Kostrzewa-Nowak D, Paine MJ, Wolf CR, Tarasiuk J (2005) The role of bioreductive activation of doxorubicin in cytotoxic activity against leukaemia HL60-sensitive cell line and its multidrug-resistant sublines. Br J Cancer 93(1):89–97
Krishnamachary N, Center MS (1993) The MRP gene associated with a non-P-glycoprotein multidrug resistance encodes a 190-kDa membrane bound glycoprotein. Cancer Res 53(16):3658–3661
Duan Z, Brakora KA, Seiden MV (2004) Inhibition of ABCB1 (MDR1) and ABCB4 (MDR3) expression by small interfering rna and reversal of paclitaxel resistance in human ovarian cancer cells. Mol Cancer Ther 3(7):833–838
Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe S, Itoh N, Shibuya M, Fukami Y (1987) Genistein, a specific inhibitor of tyrosine-specific protein kinases. J Biol Chem 262(12):5592–5595
Markovits J, Linassier C, Fosse P, Couprie J, Pierre J, Jacquemin-Sablon A, Saucier JM, Le Pecq JB, Larsen AK (1989) Inhibitory effects of the tyrosine kinase inhibitor genistein on mammalian DNA topoisomerase II. Cancer Res 49(18):5111–5117
Balabhadrapathruni S, Thomas TJ, Yurkow EJ, Amenta PS, Thomas T (2000) Effects of genistein and structurally related phytoestrogens on cell cycle kinetics and apoptosis in MDA-MB-468 human breast cancer cells. Oncol Rep 7(1):3–12
Rusin A, Gogler A, Glowala-Kosinska M, Bochenek D, Gruca A, Grynkiewicz G, Zawisza J, Szeja W, Krawczyk Z (2009) Unsaturated genistein disaccharide glycoside as a novel agent affecting microtubules. Bioorg Med Chem Lett 19(17):4939–4943
Amos LA, Schlieper D (2005) Microtubules and maps. Adv Protein Chem 71:257–298
Trock BJ, Leonessa F, Clarke R (1997) Multidrug resistance in breast cancer: a meta-analysis of MDR1/gp170 expression and its possible functional significance. J Natl Cancer Inst 89(13):917–931
Chan KF, Zhao Y, Burkett BA, Wong IL, Chow LM, Chan TH (2006) Flavonoid dimers as bivalent modulators for P-glycoprotein-based multidrug resistance: synthetic apigenin homodimers linked with defined-length poly(ethylene glycol) spacers increase drug retention and enhance chemosensitivity in resistant cancer cells. J Med Chem 49(23):6742–6759
Acknowledgments
This work was supported in part by grants from Cancer Research UK (AAA), The Zarrow Foundation, and The National Foundation for Cancer Research (AAA and RCB); European Commission grants (WKP_1/1.4.3/2/2005/74/193/385/, POIG.02.03.00-00-003/09-00—Biocentrum Ochota) and Polish Ministry of Science and Higher Education grant (PBZ-MNiI-1/1/2005) (BL) and Developmental Research Project of The University of Texas M. D. Anderson SPORE in Ovarian Cancer P50 CA-083639 (WP). We thank Dr Ron Laskey for helpful discussions, Dr Anil Sood for providing cell lines and Miss Isabella Fleminger for her contribution to making the figures.
Conflict of interest
None.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Ahmed, A.A., Goldsmith, J., Fokt, I. et al. A genistein derivative, ITB-301, induces microtubule depolymerization and mitotic arrest in multidrug-resistant ovarian cancer. Cancer Chemother Pharmacol 68, 1033–1044 (2011). https://doi.org/10.1007/s00280-011-1575-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-011-1575-2