Abstract
Interleukin-7 is a potent regulator of lymphocyte proliferation, but it inducing growth of solid tumors is few known. We study the relationship between Interleukin-7 and the regulator of the cell cycle, cyclin D1 and the mechanism of Interleukin-7 regulating cell growth in human lung cancer. We detected expression of cyclin D1 and its impact on the prognosis of lung cancer patients. Using Western blot, reverse transcriptase-PCR, Co-Immunoprecipitation, and Chromatin Immunoprecipitation, we investigated how Interleukin-7 regulated cyclin D1 in vitro and in nude mice. We found that, in lung cancer cell lines and in nude mice, Interleukin-7/Interleukin-7 receptor increased the expression of cyclin D1 and phosphorylation of c-Fos/c-Jun, induce c-Fos and c-Jun heterodimer formation, and enhanced c-Fos/c-Jun DNA-binding activity to regulate cyclin D1. In addition, lymph node metastasis, tumor stage, and cyclin D1 were the strongest predictors of survival in 100 human non-small cell lung cancer specimens analyzed. Taken together, our results provided evidence that Interleukin-7/Interleukin-7 receptor induced cyclin D1 up-regulation via c-Fos/c-Jun pathway to promote proliferation of cells in lung cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Interleukin-7 (IL-7) is a pleiotropic immune regulatory protein predominantly produced by stromal cells and by cells at the inflammatory sites [1]. IL-7 is a fundamental factor for the early development of lymphocytes and a regulator of peripheral T-cell homeostasis by modulating the expansion of peripheral T-cell populations in states of T-cell depletion [2]. IL-7 stimulates the progression of some types of lymphomas, leukemias and HIV [3–5]. IL-7 produced by some human solid tumors suggests its potential impact on the process of tumorigenesis [6, 7]. But it is unclear how IL-7 is involved in solid tumor development and progression. The studies on breast cancer describe a quantitative association between the IL-7 signaling complex and some clinicopathological parameters: there is a trend toward a higher expression of IL-7 and molecules of its signaling pathway in breast cancer patients with poor prognosis [7]. Previously, we found that the higher expression IL-7/IL-7 receptor (IL-7R) was correlated well with clinical stages, the lymph node metastasis, and short survival in human non-small cell lung cancer (NSCLC) patients [8]. Moreover, IL-7/IL-7R mRNA is detected in different tumors, such as colorectal [9], renal [10], and central nervous system cancers [11]. Following the binding of IL-7R to its ligand, a series of intracellular phosphorylation events occurred, such as the activation of the Janus kinases (JAK-1 and JAK-3), phosphoinositide 3 kinase (PI3K), and the signal transducers and activators of transcription (STAT-5) [12]. It is tempting to speculate that certain unidentified downstream gene(s) of IL-7 may have a role in tumor cell proliferation. In a previous study, we found that vascular endothelial growth factor (VEGF)-D was one of the major downstream genes of IL-7 to regulate lymph angiogenesis in lung cancer [8].
In this study, we tried to find IL-7 downstream genes of cell proliferation and study the mechanism of IL-7 inducing proliferation of tumor cells in vitro and in vivo.
Methods
Antibodies and reagents
Anti-IL-7 (mouse monoclonal, sc-365306), IL-7R (rabbit polyclonal, sc-662), cyclin C (rabbit polyclonal, sc-5610), cyclin D1 (mouse monoclonal, sc-450), cyclin E (mouse monoclonal, sc-25303), c-Jun (mouse monoclonal, sc-166540), phosphorylated c-Jun (p-c-Jun) (mouse monoclonal, sc-882), c-Fos (mouse monoclonal, sc-8047), and β-actin (mouse monoclonal, sc-81178) antibody were purchased from Santa Cruz Biotechnology (USA). 3,3′-diaminobenzidine tetrahydrochloride (DAB) was purchased from MaiXin Biotechnology (China). Separation Columns and Protein G MicroBeads were purchased from Miltenyi Biotec (Germany). Recombinant human IL-7 was purchased from Chemicon International (USA). AP-1 inhibitor (SP600125) was purchased from Calbiochem Co. (Germany).
Cell culture and siRNA
Human lung cancer cell lines A549 and LH7 (large cell carcinoma) were maintained in Dulbcco’s Modifed Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (Gibco, USA), 100 units/ml streptomycin, and 100 units/ml penicillin in a humidified 5% v/v CO2 atmosphere.
For siRNAs transfections, 105 cells were plated in 6-well dishes and serum starved for 24 h before cells were infected with siRNAs (si-cyclin D1 sc-29286 and si-IL-7R sc-35664 Santa Cruz, USA) and Lipofectamine™ 2000 reagent (Invitrogen).
In vitro cell growth assays
In 200 μl of medium, 103 cells were plated in a 96-well plate and incubated for 24, 48, and 72 h. Added to each well was 20 μl of 5 mg/ml MTT (3-[4, 5–2-yl]- 2,5-diphenyl tetrazolium bromide; Sigma). After a 4 h incubation, the medium containing MTT was removed and replaced with 150 μl of dimethyl sulfoxide (DMSO; Sigma) for further incubation for 10 min until the formazan was dissolved. The optical density (OD) value of each well was measured using a microplate reader (Spectra Thermo, USA) with a test wavelength of 490 nm. The wells containing only the medium were used as controls. The proliferation curve was plotted.
Flow cytometry (FACS, fluorescence-activated cell sorting)
A549 and LH7 cells were starved for 48 h and then stimulated with serum for the indicated time periods. Wherever indicated, 24 h post-transfected cells were subjected to serum-starvation for another 24 h. Flow cytometry of cells was done as described [13].
Western blot analysis
Total protein was extracted with lysis buffer (150 mM NaCl, 1% v/v NP-40, 0.1% v/v SDS, 2 μg/ml aprotinin, 1 mM PMSF), and 60 μg of protein lysates were separated on a 12% v/v SDS–polyacrylamide electrophoresis gel, transferred to Polyvinylidene Fluoride (PVDF) membranes. Proteins were visualized with horse-radish peroxidase-conjugated goat anti-rabbit and anti-mouse IgG (Zhongshan, Beijing, China) followed by DAB. Subsequently, densitometric analyses of the bands were performed.
RNA isolation and reverse transcriptase-PCR
Total RNA was isolated from cells in the logarithmic growth phase using TRIZOL (Invitrogen, USA). The PCR primers are listed in Table 1.
Co-immunoprecipitation (CoIP)
The protein was extracted with cellular lysis buffer. Equal amounts of protein were incubated with c-Jun-specific antibody immobilized onto protein G-bead for 1 h at 4°C with gentle rotation. Beads were washed extensively with lysis buffer, boiled, and micro centrifuged. Proteins were detected with c-Fos antibody by Western blot analysis.
Chromatin immunoprecipitation (CHIP)
We performed the ChIP assay according to the instructions of the ChIP assay kit (Upstate, USA). The procedure included DNA–protein cross-linking in chromatin, shearing DNA into smaller fragments, immunoprecipitation with anti-c-Jun antibody (negative control with normal rabbit IgG), followed by PCR identification of associated DNA sequences. The PCR primer sequences designed according to cyclin D1 promoter are the following: forward (5′-CAG TCC CAG GGC AAA TTC TA-3′) and reverse (5′-TTA ACC GGG AGA AAC ACA CC-3′). The PCR product is 200 bp. The positive control group used total DNA.
Experiment on nude mice with xenograft tumors
Forty male BALB/c nude (nu/nu) mice (Chinese Academy of Medical Sciences) were used, 4 weeks of age at the start of the experiment (17–18 g). The animals were maintained under specific pathogen-free conditions. A549 cells (2 × 106 per mouse) were injected subcutaneous in 100 μl serum-free medium. All of the animals were randomly divided into four groups when the maximum diameter of the tumor reached 5–7 mm. There were ten mice in each group. In the Group A mice, 100 μl of recombinant human IL-7 (20 μg/ml/kg) in of serum-free medium was directly injected into the tumor once every 2 days. The group B mice were injected with 100 μl of IL-7R-specific antibody (sc-662, 100 μg/ml/kg) in of serum-free medium once every 2 days. The group C mice were administered with SP600125 (25 μmol/ml/kg) in of serum-free medium once every 2 days. The group D comprised control mice that were injected with 100 μl serum-free medium once every 2 days. The tumor volume was excised once a week. The tumor volume was calculated by the formula: volume = (width)2 × length/2. Data are presented as the mean ± SD. The growth curve of the tumor was drawn. Mice were killed by depleting 6 weeks after the first treatment, and tumors were excised and weighed. The fraction of every tumor from every animal the tumor tissue was frozen instantly for RT-PCR, western blot, CoIP, and CHIP detecting.
Patients and specimens
A total of 100 cases of NSCLC were obtained from the January 1, 1980 to the December 31, 2005 at the First Affiliated Hospital of China Medical University, Shenyang, China. The tumor tissues in this study were from patients who had NSCLC proved by pathological diagnosis without distant metastasis. None of the 100 cases had received radiation therapy or chemotherapy before surgery. The TNM staging system of the UICC (1997) was used to classify the specimens. In these cases, 57 showed lymph node metastasis (Table 2). The survival time was calculated from the operation day to death via the evaluation of recurrence and metastasis or until the last follow-up date (December 2006). The following-up of the surviving patients averaged 23.09 months and ranged from 1 to 117 months. The study has been approved by the Hospitals’ Ethical Review Committee.
Immunohistochemistry
Four-micron-thick sections were prepared from the paraffin-embedded formalin-fixed tissues. Immunostaining was performed by the streptavidin-peroxidase (S-P) method (Ultrasensitive™ MaiXin, Fuzhou, China). The primary antibodies were anti-IL-7, anti-IL-7R, and anti-cyclin D1 (1:100, 1:100, 1:150) antibodies. The peroxidase reaction was developed with DAB. For negative control, the primary antibodies were replaced by non-immune serum.
We counted 200 tumor cells and calculated the percentage of positively stained cells. The proportion of cells exhibiting IL-7, IL-7R, and cyclin D1 expression was categorized as follows: 0, absent; 1, 1–25%; 2, 26–50%; 3, 51–75%; 4, more than 75%. The staining intensity was categorized as follows: 1, weak; 2, moderate; 3, strong. The proportion and intensity scores were then multiplied to obtain a total score. A score <3 was considered low expression.
Statistical analysis
The statistical package SPSS13.0 (SPSS incorporated, Chicago) was used for all analysis. The Chi-square test, Kaplan–Meier curves, t test, log-rank, and Cox regression multivariate analysis were used to analyze data. Values of P < 0.05 were considered statistically significant.
Results
IL-7/IL-7R promote the proliferation of cells in lung cancer cell lines
Using MTT approach, we found that lung cancer A549 and LH7 cells proliferation (P < 0.05) was promoted after incubation with the IL-7 (20 ng/ml) and inhibited with siRNA blocking IL-7R (Fig. 1). Thus, IL-7 could promote the proliferative ability of A549 cell by IL-7R.
IL-7/IL-7R accelerate G1/S-phase progression in lung cancer cell lines
The FACS analysis clearly indicated that IL-7 could promote G1/S-phase transition in A549 and LH7 cells. However, siRNA against IL-7R inhibited G1/S-phase progression in A549 and LH7 cells (Fig. 2). It suggested that IL-7 could facilitate cells into S-phase of cell cycle via IL-7R.
IL-7/IL-7R regulate the cell cycle by cyclin D1 in lung cancer cell lines
Cyclin C, D1, E are essential for cellular progression through the G1 phase of the cell cycle and initiation of DNA replication. We explored if IL-7 accelerated G1/S-phase progression of cell cycle via three regulators. We found that treatment of A549 and LH7 cells with recombinant human IL-7 increased the expression of cyclin D1 mRNA and protein (Fig. 3), while the expression of cyclin C and E mRNA and protein were not affected in A549 and LH7 cells after IL-7 stimulation. Then, we detected the expression of cyclin C, D1, and E mRNA and protein of A549 and LH7 cells after blocking IL-7R with siRNA. Blockage of IL-7R decreased the expression of cyclin D1 mRNA and protein, which did not affected the expression of cyclin C and E mRNA and protein (Fig. 3). It suggested that IL-7 could up-regulate cyclin D1 via IL-7R. It suggested that IL-7 could up-regulate via cyclin D1.
IL-7 up-regulates cyclin D1 in lung cancer cell lines. The RT-PCR and Western blot analysis showing induction expression of cyclin D1 mRNA and protein after Il-7 stimulation and reduction expression of cyclin D1 mRNA and protein after blocking IL-7R with siRNA IL-7R in A549 (a, b) and LH7 (c, d) cells. However, after Il-7 stimulation or blocking IL-7R with siRNA, IL-7R did not affect the expression of cyclin C and cyclin E protein and mRNA in A549 (a, b) and LH7 (c, d) cells
To further examine whether IL-7 up-regulated expression of cyclin D1 to accelerate G1/S-phase progression in lung cancer cell lines, we observed the change cell cycle of A549 and LH7 with siRNA inhibiting cyclin D1. The result of FACS showed that the change cell cycle was similar with blocking IL-7R with siRNA (Fig. 4). It suggested that IL-7/IL-7R could up-regulate S-phase entry via cyclin D1.
IL-7/IL-7R induce cyclin D1 via c-Fos/c-Jun pathway
To investigate how IL-7/IL-7R regulated cyclin D1, we checked the expression of AP-1 protein in A549 and LH7 cells. Incubation of the cells with IL-7 increased the expression of c-Fos, c-Jun, and p-c-Jun protein, while blocking IL-7R with siRNA decreased the expressions of c-Fos, c-Jun, and p-c-Jun (Fig. 5).
IL-7 up-regulates the expression and phosphorylation of c-Fos, c-Jun in lung cancer cell lines. Western blot analysis showing up-regulation of c-Fos, c-Jun, and p-c-Jun proteins in A549 (a) and LH7 (b) cells treated with IL-7, and blocking IL-7R with siRNA IL-7R down-regulating the expression of c-Fos, c-Jun, and p-c-Jun proteins
To confirm whether IL-7/IL-7R up-regulated cyclin D1 through activation of c-Fos/c-Jun, then we inhibited the activity of c-Fos/c-Jun with a specific AP-1 inhibitor SP600125. RT-PCR and Western blotting analyses showed that the expressions of cyclin D1 mRNA and protein in A549 and LH7 cells were decreased significantly after treatment with SP600125, while decreased expression of cyclin D1 was not affected by the IL-7 (Fig. 6). These results imply that IL-7/IL-7R up-regulate cyclin D1 via c-Fos/c-Jun.
IL-7/IL-7R enhance the DNA-binding activity of AP-1 to the promoter of cyclin D1
AP-1 is a transcription factor that binds to the promotor of specific target gene to regulate its transcription. c-Fos and c-Jun could form heterodimer to enhance its activity [14]. We had found that IL-7/IL-7R promote the formation of c-Fos/c-Jun heterodimer in previous study [8]. To examine whether AP-1 binds to cyclin D1 promoter, we first analyzed promoter sequence of cyclin D1 gene and identified AP-1 binding sites (consensus sequence 5′-TGAG/CTCA-3′), also known as TPA-responsive elements (TREs). We then performed CHIP assay. CHIP analysis demonstrated that AP-1 could bind to cyclin D1 promoter. Then, we detected AP-1 DNA-binding activity in A549 cell after incubation with the IL-7 or transfection with siRNA IL-7R. IL-7 enhanced AP-1 binding to cyclin D1 promoter, while blocking IL-7R reduced AP-1 binding to cyclin D1 promoter (Fig. 7).
IL-7 enhances the DNA-binding activity of AP-1 to the Promoter of Cyclin D1. The CHIP analysis showing that AP-1 (c-Fos/c-Jun) could bind with cyclin D1 promoter, and the activity of binding was enhanced by IL-7 in A549 cell line. However, inhibiting IL-7R with siRNA IL-7R decreased the activity of binding AP-1 with cyclin D1 promoter
IL-7/IL-7R induce cyclin D1 via AP-1 to promote cell growth in vivo
To examine whether IL-7 regulated expression of cyclin D1 could be observed in vivo, we next study experiment on nude mice with xenograft tumors, respectively, with the IL-7, sc-662, and SP600125 treatment. We excised tumor volume once a week and found IL-7 promotes tumors growth, and blocking IL-7R or AP-1 inhibited tumors growth (Fig. 8), suggesting that IL-7 could induce tumor cells growth in vivo. Then, we found that IL-7 increased the expression of cyclin D1 protein and mRNA levels in xenograft tumors, and blocking IL-7R or AP-1 decreased their expression levels (Fig. 9a, b), indicating that IL-7 could up-regulate cyclin D1 via IL-7R in vivo. IL-7 increased the expression of c-Fos and c-Jun, and p-c-Jun protein, while blocking IL-7R or AP-1 decreased the expressions of c-Fos, c-Jun, and p-c-Jun (Fig. 9c), implying that IL-7/IL-7R up-regulate cyclin D1 via c-Fos/c-Jun in vivo. Using CoIP approach, we found that c-Fos/c-Jun heterodimer was increased after incubation with the IL-7, and blocking IL-7R or AP-1 decreased the dimer formation (Fig. 9d), suggesting that IL-7/IL-7R could promote c-Fos and c-Jun to form heterodimer. CHIP analysis demonstrated that AP-1 could bind to cyclin D1 promoter. Then, we detected AP-1 DNA-binding activity in xenograft tumors. IL-7 enhanced AP-1 binding to cyclin D1 promoter, while blocking IL-7R reduced AP-1 binding to cyclin D1 promoter (Fig. 9e) in vivo. These data implied that IL-7/IL-7R up-regulate cyclin D1 via c-Fos/c-Jun to tumors growth.
The RT-PCR and Western blot analysis showing induction of cyclin D1 mRNA and protein after Il-7 stimulation and reduction in cyclin D1 mRNA and protein after blocking IL-7R or inhibiting AP-1 in nude mice (a, b). Western blot analysis showing up-regulation of c-Fos, c-Jun, and p-c-Jun proteins in nude mice treated with IL-7, and blocking IL-7R with IL-7R-specific antibody (sc-662) or inactivating AP-1 with AP-1 inhibitor (SP600125) down-regulating the expression of c-Fos, c-Jun, and p-c-Jun proteins (c). IL-7 could induce the formation of AP-1 (c-Fos/c-Jun) heterodimmer via IL-7R with CoIP method in vivo (d). The CHIP analysis showing that AP-1 (c-Fos/c-Jun) could bind with cyclin D1 promoter, and the activity of binding were enhanced by IL-7 in xenograft tumors (e)
Expression of cyclin D1 correlate with IL-7/IL-7R level and NSCLC patient survival
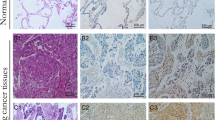
Immunohistochemical analysis of 100 NSCLC specimens revealed that the expression of cyclin D1 was significantly associated with the expression of IL-7 and IL-7R (Table 2; Fig. 10).
The expressions of cyclin D1 in NSCLC correlates with the level of IL-7 and IL-7R. Immunohistochemical straining of consecutive serial sections of NSCLC tissues. Brown-yellow particles of IL-7 (a), IL-7R (b), and cyclin D1 (c) were observed in cancer cells (relationship between cyclin D1 expression with IL-7 (P = 0.001, R = 11.063) and with IL-7R (P = 0.025, R = 5.365); 400 × magnification)
Clinically, the expression levels of cyclin D1 in NSCLC did not correlated with sex, age, differentiation, clinical stages, and lymph node metastasis. Patients with low expression of cyclin D1 had a statistically significantly longer survival than those with high expression of cyclin D1 (median survival = 10 ± 4.1 months; 95% CI 1.9–18.1 months; P = 0.000; Fig. 11). Previously, we had found patients with low expression of IL-7 and IL-7R survived longer. Univariate analysis showed that lymph node metastasis, tumor stage, IL-7, IL-7R, and cyclin D1 could affect the survival. In the Cox regression multivariate analysis, lymph node metastasis, tumor stage, and cyclin D1 were the strongest predictors of survival (Table 3). These results provide a possible explanation that IL-7, IL-7R, and cyclin D1 high expression patients were more likely to have poor prognosis, possibly resulting from IL-7/IL-7R-mediated cell proliferation via up-regulation of cyclin D1 in human NSCLC.
Survival of NSCLC patients correlates with the expression of cyclin D1. Kaplan–Meier survival plots for patients with NSCLC, grouped according to cyclin D1 protein expression. Correlation between overall survival of patients with cyclin D1 expression was found to be statistically significant (P = 0.000). All patients alive at their last follow-up are indicated by tick marks on the plot
Discussion
Previous evidences had suggested the important role of IL-7 in the pathogenesis and progression of lymphomas [3, 15]. In breast cancer cell lines, IL-7 could induce the growth of cells, while this effect involved PI3K and Jak3 [16].
In the present study, we found IL-7 promoted cell growth. Then, we detected the effect of IL-7 to cell cycle. The result suggested that IL-7 could regulate G1/S stage of cell cycle. Cyclin C, D1, and E were important members of the G1 cyclin family involved in the regulation of the G1/S transition of the cell cycle [17]. We examined whether IL-7 was interrelated with cyclin C, D1, or E in lung cancer cell. We obtained that recombinant human IL-7 increased the expression of cyclin D1 mRNA and protein in lung cancer cell lines, and blocking IL-7R with siRNA could abolish the role of IL-7 on cyclin D1. But the recombinant human IL-7 and blocking IL-7R with siRNA did not affect cyclin C and E. The cyclin D1 was frequently overexpressed in a wide range of cancers. The nuclear accumulation of cyclin D1 induced uncontrolled proliferation in normal human cells, which could facilitate the development of invasive cancer [18]. In addition, the result of FACS showed that the change cell cycle was similar with blocking IL-7R with siRNA. It suggested that IL-7/IL-7R could up-regulate S-phase entry via cyclin D1.
The cyclin D1 expression was under complex regulation and markedly influenced by the activator protein-1 (AP-1), NF-kB, and b-catenin/T-cell factor (TCF) signaling pathways [19–21]. A number of compounds targeting these signaling pathways could indirectly attenuate the cyclin D1 expression to mediate cell cycle arrest.
Previously, we found IL-7 induced c-Fos, c-Jun expression, and phosphorylation, promoted c-Fos and c-Jun heterodimer formation, and enhanced the activity of c-Fos/c-Jun. AP-1 was a sequence-specific transcription factor composed of homodimers of the Jun family (c-Jun, Jun D, and Jun B) or heterodimers of the Jun family members with any of the Fos family members (c-Fos, Fos B, Fra1, and Fra2). AP-1 had long been associated with proliferation. AP-1 directed the expression of a critical target gene or genes, in response to cytokines, stress, and mitogenic signals. A common feature of all these proteins was the evolutionarily conserved bZIP domain, the collective term for a basic DNA-binding domain combined with a leucine zipper region. The leucine zipper was responsible for dimerization, which was a prerequisite for DNA binding mediated by the basic domain. AP-1 had long been associated with proliferation [22]. We found that AP-1 can bind to cyclin D1 promoter. Consistent with our finding, it had been shown that berberine inhibited cyclin D1 expression via suppressing binding of AP-1 transcription factors to CCND1 AP-1 motif [23].
In nude mice xenograft tumors, we obtained IL-7 could induce tumor growth, up-regulate the expressions of c-Fos, c-Jun, p-c-Jun, and cyclin D1, and promote AP-1 binding activity. This result was consistent with it in lung cancer cell lines.
Immunohistochemical analysis of 100 NSCLC specimens revealed the positive expression of IL-7, IL-7R, and cyclin D1 in lung cancer cells. It had been shown that a few colon cancer cells could produce IL-7, breast cancer tissues expressed IL-7 and IL-7R, and IL-7R positively correlated with tumors metastasized to the regional lymph nodes [24]. In previous study, we found that the high expression of IL-7 and IL-7R correlated well with lymph node metastasis and poor prognosis [8]. In this study, we further evaluated expression of cyclin D1 in 100 of NSCLC tissues. The expression of cyclin D1 was positive in lung cancer cells. Interestingly, we had found that the high expression of cyclin D1 correlated well with high expression of IL-7 and IL-7R and shorter survival rate of patients. Cox regression multivariate analysis showed lymph node metastasis, tumor stage, and cyclin D1 were the strongest predictors of survival.
Conclusion
In this study demonstrated that, in vivo and in vitro, IL-7 and its receptor IL-7R, were able to induce cyclin D1 gene expression via AP1 (c-Fos/c-Jun)-dependent pathway to promote cell growth in human lung cancer. In situ analysis of human, NSCLC revealed that overexpression of IL-7/IL-7R and cyclin D1 played an important role in lung cancer development. We inhibited IL-7R to reduce cell proliferation in lung cancer. Thus, targeting IL-7/IL-7R may potentiate new therapeutic strategy against tumors.
References
Appasamy PM (1999) Biological and clinical implications of interleukin-7 and lymphopoiesis. Cytokines Cell Mol Ther 5:25–39
Fry TJ, Mackall CL (2001) Interleukin-7: master regulator of peripheral T-cell homeostasis? Trends Immunol 22:564–571
Yamanaka K, Clark R, Rich B, Dowgiert R, Hirahara K, Hurwitz D et al (2006) Skin-derived interleukin-7 contributes to the proliferation of lymphocytes in cutaneous T-cell lymphoma. Blood 107:2440–2445
González-García S, García-Peydró M, Martín-Gayo E, Ballestar E, Esteller M, Bornstein R et al (2009) CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7R {alpha} gene expression in early human thymopoiesis and leukemia. J Exp Med 206:779–791
Rajasuriar R, Booth D, Solomon A, Chua K, Spelman T, Gouillou M et al (2010) Biological determinants of immune reconstitution in HIV-infected patients receiving antiretroviral therapy: the role of interleukin 7 and interleukin 7 receptor α and microbial translocation. J Infect Dis 202:1254–1264
Al-Rawi MA, Mansel RE, Jiang WG (2003) Interleukin-7 (IL-7) and IL-7 receptor (IL-7R) signalling complex in human solid tumours. Histol Histopathol 18:911–923
Al-Rawi MA, Rmali K, Watkins G, Mansel RE, Jiang WG (2004) Aberrant expression of interleukin-7 (IL-7) and its signalling complex in human breast cancer. Eur J Cancer 40:494–502
Ming J, Zhang Q, Qiu X, Wang E (2009) Interleukin 7/interleukin 7 receptor induce c-Fos/c-Jun-dependent vascular endothelial growth factor-D up-regulation: a mechanism of Lymphangiogenesis in lung cancer. Eur J Cancer 45:866–873
Maeurer MJ, Walter W, Martin D, Zitvogel L, Elder E, Storkus W et al (1997) Interleukin-7 (IL-7) in colorectal cancer: IL-7 is produced by tissues from colorectal cancer and promotes preferential expansion of tumour infiltrating lymphocytes. Scand J Immunol 45:182–192
Trinder P, Seitzer U, Gerdes J, Seliger B, Maeurer M (1999) Constitutive and IFN-gamma regulated expression of IL-7 and IL-15 in human renal cell cancer. Int J Oncol 14:23–31
Cosenza L, Gorgun G, Urbano A, Foss F (2002) Interleukin-7 receptor expression and activation in non haematopoietic neoplastic cell lines. Cell Signal 14:317–325
Foxwell BMJ, Beadling C, Guschin D, Kerr I, Cantrell D (1995) Interleukin-7 can induce the activation of JAK-1, JAK-3 and STAT-5 proteins in murine T cells. Eur J Immunol 25:3041–3046
Mukherji A, Janbandhu VC, Kumar V (2007) HBx-dependent cell cycle deregulation involves interaction with cyclin E/A-cdk2 complex and destabilization of p27Kip1. Biochem J 401:247–256
Hess J, Angel P, Schorpp-Kistner M (2004) AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 117:5965–5973
Takakuwa T, Nomura S, Matsuzuka F, Inoue H, Aozasa K (2000) Expression of interleukin-7 and its receptor in thyroid lymphoma. Lab Invest 80:1483–1490
Al-Rawi A, Rmali K, Mansel RE, Jiang WG (2004) Interleukin 7 induces the growth of breast cancer cells through a wortmannin-sensitive pathway. Br J Surg 91:61–68
Matsushime H, Roussel MF, Ashmun RA, Sherr CJ (1991) Colonystimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell 65:701–713
Gautschi O, Ratschiller D, Gugger M, Betticher DC, Heighway J (2007) Cyclin D1 in non-small cell lung cancer: a key driver of malignant transformation. Lung Cancer 55:1–14
Eferl R, Wagner EF (2003) AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3:859–868
Joyce D, Bouzahzah B, Fu M, Albanese C, D’Amico M, Steer J et al (1999) Integration of Rac-dependent regulation of cyclin D1 transcription through a nuclear factor-kappaB-dependent pathway. J Biol Chem 274:25245–25249
Natsume H, Sasaki S, Kitagawa M, Kashiwabara Y, Matsushita A, Nakano K et al (2003) Beta-catenin/Tcf-1-mediated transactivation of cyclin D1 promoter is negatively regulated by thyroid hormone. Biochem Biophys Res Commun 309:408–413
Shaulian E, Karin M (2001) AP-1 in cell proliferation and survival. Oncogene 20:2390–2400
Luo Y, Hao Y, Shi TP, Deng WW, Li N (2008) Berberine inhibits cyclin D1 expression via suppressed binding of AP-1 transcription factors to CCND1 AP-1 motif. Acta Pharmacol Sin 29:628–633
Al-Rawi M, Mansel R, Jiang W (2002) Interleukin-7 (IL-7) and IL-7 receptor expression in breast cancer. Breast Cancer Res Treat 76(Suppl 1):144
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 30972967) and Specialized Research Fund for the Doctoral Program of Higher Education (No. 20092104110018).
Conflict of interest
The authors declare that they have no competing interests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Ming, J., Jiang, G., Zhang, Q. et al. Interleukin-7 up-regulates cyclin D1 via activator protein-1 to promote proliferation of cell in lung cancer. Cancer Immunol Immunother 61, 79–88 (2012). https://doi.org/10.1007/s00262-011-1078-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-011-1078-3