Abstract
Purpose
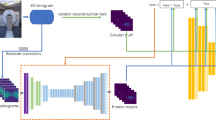
The PET scanners with long axial field of view (AFOV) having ~ 20 times higher sensitivity than conventional scanners provide new opportunities for enhanced parametric imaging but suffer from the dramatically increased volume and complexity of dynamic data. This study reconstructed a high-quality direct Patlak Ki image from five-frame sinograms without input function by a deep learning framework based on DeepPET to explore the potential of artificial intelligence reducing the acquisition time and the dependence of input function in parametric imaging.
Methods
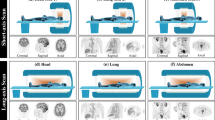
This study was implemented on a large AFOV PET/CT scanner (Biograph Vision Quadra) and twenty patients were recruited with 18F-fluorodeoxyglucose (18F-FDG) dynamic scans. During training and testing of the proposed deep learning framework, the last five-frame (25 min, 40–65 min post-injection) sinograms were set as input and the reconstructed Patlak Ki images by a nested EM algorithm on the vendor were set as ground truth. To evaluate the image quality of predicted Ki images, mean square error (MSE), peak signal-to-noise ratio (PSNR), and structural similarity index measure (SSIM) were calculated. Meanwhile, a linear regression process was applied between predicted and true Ki means on avid malignant lesions and tumor volume of interests (VOIs).
Results
In the testing phase, the proposed method achieved excellent MSE of less than 0.03%, high SSIM, and PSNR of ~ 0.98 and ~ 38 dB, respectively. Moreover, there was a high correlation (DeepPET: \({R}^{2}\)= 0.73, self-attention DeepPET: \({R}^{2}\)=0.82) between predicted Ki and traditionally reconstructed Patlak Ki means over eleven lesions.
Conclusions
The results show that the deep learning–based method produced high-quality parametric images from small frames of projection data without input function. It has much potential to address the dilemma of the long scan time and dependency on input function that still hamper the clinical translation of dynamic PET.













Similar content being viewed by others
References
Muehllehner G, Karp JS. Positron emission tomography. Phys Med Biol. 2006;51(13);R117–R137. https://doi.org/10.1088/0031-9155/51/13/R08.
Beyer T, Bidaut L, Dickson J, Kachelriess M, Kiessling F, Leitgeb R, et al. What scans we will read: imaging instrumentation trends in clinical oncology. Cancer Imaging. 2020;20:1–38.
Karakatsanis NA, Lodge MA, Tahari AK, Zhou Y, Wahl RL, Rahmim A. Dynamic whole-body PET parametric imaging: I Concept, acquisition protocol optimization and clinical application. Phys Med Biol. 2013;58:7391–418.
Huang SC. Anatomy of SUV. Nucl Med Biol. 2000;27:643–6.
Dimitrakopoulou-Strauss A, Pan L, Sachpekidis C. Kinetic modeling and parametric imaging with dynamic PET for oncological applications: general considerations, current clinical applications, and future perspectives. Eur J Nucl Med Mol Imaging. 2021;48:21–39.
Carson RE. Tracer kinetic modeling in PET. Positron Emiss Tomogr. 2006;127–59.
Tsoumpas C, Turkheimer FE, Thielemans K. A survey of approaches for direct parametric image reconstruction in emission tomography. Med Phys. 2008;35:3963–71.
Tsoumpas C, Turkheimer FE, Thielemans K. Study of direct and indirect parametric estimation methods of linear models in dynamic positron emission tomography. Med Phys. 2008;35:1299–309.
Karakatsanis NA, Casey ME, Lodge MA, Rahmim A, Zaidi H. Whole-body direct 4D parametric PET imaging employing nested generalized Patlak expectation-maximization reconstruction. Phys Med Biol. 2016;61:5456–85. https://doi.org/10.1088/0031-9155/61/15/5456.
Rahmim A, Tang J, Zaidi H. Four-dimensional (4D) image reconstruction strategies in dynamic PET: beyond conventional independent frame reconstruction. Med Phys. 2009;36:3654–70.
Reader AJ, Verhaeghe J. 4D image reconstruction for emission tomography. Phys Med Biol. 2014;59:R371-418.
Cheng X, Bayer C, Maftei CA, Astner ST, Vaupel P, Ziegler SI, et al. Preclinical evaluation of parametric image reconstruction of [18F]FMISO PET: correlation with ex vivo immunohistochemistry. Phys Med Biol. 2014;59:347–62.
Hu J, Panin V, Smith AM, Spottiswoode B, Shah V, CA von Gall C, et al. Design and implementation of automated clinical whole body parametric PET with continuous bed motion. IEEE Trans Radiat Plasma Med Sci. 2020;4:696–707
Dias AH, Pedersen MF, Danielsen H, Munk OL, Gormsen LC. Clinical feasibility and impact of fully automated multiparametric PET imaging using direct Patlak reconstruction: evaluation of 103 dynamic whole-body 18F-FDG PET/CT scans. Eur J Nucl Med Mol Imaging. 2021;48:837–50.
Cherry SR, Jones T, Karp JS, Qi J, Moses WW, Badawi RD. Total-body PET: maximizing sensitivity to create new opportunities for clinical research and patient care. J Nucl Med. 2018;59:3–12.
Zhang X, Zhou J, Cherry SR, Badawi RD, Qi J. Quantitative image reconstruction for total-body PET imaging using the 2-meter long EXPLORER scanner. Phys Med Biol. 2017;62:2465–85.
Van der Weerdt AP, Klein LJ, Visser CA, Visser FC, Lammertsma AA. Use of arterialised venous instead of arterial blood for measurement of myocardial glucose metabolism during euglycaemic-hyperinsulinaemic clamping. Eur J Nucl Med. 2002;29:663–9.
Chen K, Bandy D, Reiman E, Huang SC, Lawson M, Feng D, et al. Noninvasive quantification of the cerebral metabolic rate for glucose using positron emission tomography, 18F-fluoro-2-deoxyglucose, the Patlak method, and an image-derived input function. J Cereb Blood Flow Metab. 1998;18:716–23.
Wu HM, Hoh CK, Choi Y, Schelbert HR, Hawkins RA, Phelps ME, et al. Factor analysis for extraction of blood time-activity curves in dynamic FDG-PET studies. J Nucl Med. 1995;36:1714–22.
Croteau E, Lavallée É, Labbe SM, Hubert L, Pifferi F, Rousseau JA, et al. Image-derived input function in dynamic human PET/CT: methodology and validation with 11C-acetate and 18F-fluorothioheptadecanoic acid in muscle and 18F-fluorodeoxyglucose in brain. Eur J Nucl Med Mol Imaging. 2010;37:1539–50.
Sari H, Erlandsson K, Law I, Larsson HBW, Ourselin S, Arridge S, et al. Estimation of an image derived input function with MR-defined carotid arteries in FDG-PET human studies using a novel partial volume correction method. J Cereb Blood Flow Metab. 2017;37:1398–409.
Sundar LKS, Muzik O, Rischka L, Hahn A, Rausch I, Lanzenberger R, et al. Towards quantitative [18F]FDG-PET/MRI of the brain: automated MR-driven calculation of an image-derived input function for the non-invasive determination of cerebral glucose metabolic rates. J Cereb Blood Flow Metab. 2019;39:1516–30.
Roccia E, Mikhno A, Ogden RT, Mann JJ, Laine AF, Angelini ED, et al. Quantifying brain [18F]FDG uptake noninvasively by combining medical health records and dynamic PET imaging data. IEEE J Biomed Heal Informatics. 2019;23:2576–82.
Kuttner S, Wickstrm KK, Kalda G, Dorraji SE, Martin-Armas M, Oteiza A, et al. Machine learning derived input-function in a dynamic 18F-FDG PET study of mice. Biomed Phys Eng Express. 2020;6(1):015020. https://doi.org/10.1088/2057-1976/ab6496.
Feng T, Zhao Y, Shi H, Li H, Zhang X, Wang G, et al. Total-body quantitative parametric imaging of early kinetics of 18 F-FDG. J Nucl Med. 2021;62:738–44.
Badawi RD, Shi H, Hu P, Chen S, Xu T, Price PM, et al. First human imaging studies with the explorer total-body PET scanner. J Nucl Med. 2019;60:299–303.
Spencer BA, Berg E, Schmall JP, Omidvari N, Leung EK, Abdelhafez YG, et al. Performance evaluation of the uEXPLORER total-body PET/CT scanner based on NEMA NU 2–2018 with additional tests to characterize PET scanners with a long axial field of view. J Nucl Med. 2021;62:861–70.
Pantel AR, Viswanath V, Daube-Witherspoon ME, Dubroff JG, Muehllehner G, Parma MJ, et al. PennPET explorer: human imaging on a whole-body imager. J Nucl Med. 2020;61:144–51.
Prenosil GA, Sari H, Fürstner M, Afshar-Oromieh A, Shi K, Rominger A, et al. Performance characteristics of the Biograph Vision Quadra PET/CT system with long axial field of view using the NEMA NU 2–2018 Standard. J Nucl Med. 2021;121:261972.
Zhang X, Xie Z, Berg E, Judenhofer MS, Liu W, Xu T, et al. Total-body dynamic reconstruction and parametric imaging on the uEXPLORER. J Nucl Med. 2020;61:285–91.
Alberts I, Hünermund JN, Prenosil G, Mingels C, Bohn KP, Viscione M, et al. Clinical performance of long axial field of view PET/CT: a head-to-head intra-individual comparison of the Biograph Vision Quadra with the Biograph Vision PET/CT. Eur J Nucl Med Mol Imaging. 2021;48:2395–404.
Sari H, Mingels C, Alberts I, Hu J, Buesser D, Shah V, et al. First results on kinetic modelling and parametric imaging of dynamic 18F-FDG datasets from a long axial FOV PET scanner in oncological patients. Eur J Nucl Med Mol Imaging. 2022. https://doi.org/10.1007/s00259-021-05623-6.
Viswanath V, Sari H, Pantel AR, Conti M, Daube-Witherspoon ME, Mingels C, et al. Abbreviated scan protocols to capture 18F-FDG kinetics for long axial FOV PET scanners. Eur J Nucl Med Mol Imaging. 2022. https://doi.org/10.1007/s00259-022-05747-3.
Xu J, Gong E, Pauly J, Zaharchuk G. 200x Low-dose PET reconstruction using deep learning. 2017. http://arxiv.org/abs/1712.04119. Accessed 8 Dec 2017.
Cui J, Gong K, Guo N, Wu C, Kim K, Liu H, et al. Populational and individual information based PET image denoising using conditional unsupervised learning. Phys Med Biol. 2021;66(15). https://doi.org/10.1088/1361-6560/ac108e.
Lu W, Onofrey JA, Lu Y, Shi L, Ma T, Liu Y, et al. An investigation of quantitative accuracy for deep learning based denoising in oncological PET. Phys Med Biol. 2019;64(16):165019.
Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, et al. PET image denoising using unsupervised deep learning. Eur J Nucl Med Mol Imaging. 2019;46:2780–9.
Niyas S, Pawan SJ, Kumar MA, Rajan J. Medical image segmentation using 3D convolutional neural networks: a review. 2021. http://arxiv.org/abs/2108.08467. Accessed 9 Apr 2022.
Gu Z, Cheng J, Fu H, Zhou K, Hao H, Zhao Y, et al. CE-Net: context encoder network for 2D medical image segmentation. IEEE Trans Med Imaging. 2019;38:2281–92.
Gong K, Guan J, Kim K, Zhang X, Yang J, Seo Y, et al. Iterative PET image reconstruction using convolutional neural network representation. IEEE Trans Med Imaging. 2019;38:675–85.
Gong K, Yang J, Kim K, El Fakhri G, Seo Y, Li Q. Attenuation correction for brain PET imaging using deep neural network based on Dixon and ZTE MR images. Phys Med Biol. 2018;63.
Sun Y, Xu W, Zhang J, Xiong J, Gui G. Super-resolution imaging using convolutional neural networks. Lect Notes Electr Eng. 2020;516:59–66.
Hu Z, Xue H, Zhang Q, Gao J, Zhang N, Zou S, et al. DPIR-Net: direct PET image reconstruction based on the Wasserstein Generative Adversarial Network. IEEE Trans Radiat Plasma Med Sci. 2021;5:35–43.
Häggström I, Schmidtlein CR, Campanella G, Fuchs TJ. DeepPET: a deep encoder–decoder network for directly solving the PET image reconstruction inverse problem. Med Image Anal. 2019;54:253–62.
Wang G, Qi J. Direct estimation of kinetic parametric images for dynamic PET. Theranostics. 2013;3:802–15.
Scipioni M, Giorgetti A, Della Latta D, Fucci S, Positano V, Landini L, et al. Accelerated PET kinetic maps estimation by analytic fitting method. Comput Biol Med. 2018;99:221–35. https://doi.org/10.1016/j.compbiomed.2018.06.015.
Reader AJ, Corda G, Mehranian A, da Costa-Luis C, Ellis S, Schnabel JA. Deep learning for PET image reconstruction. IEEE Trans Radiat Plasma Med Sci. 2020;5:1–25.
Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. Adv Neural Inf Process Syst. 2017;5999–6009.
Li M, Hsu W, Xie X, Cong J, Gao W. SACNN: Self-Attention Convolutional Neural Network for low-dose CT denoising with self-supervised perceptual loss network. IEEE Trans Med Imaging. 2020;39:2289–301.
Fu J, Liu J, Tian H, Li Y, Bao Y, Fang Z, et al. Dual attention network for scene segmentation. Proc IEEE Conf Comput Vis Pattern Recognit. (CVPR). 2017;299–307.
Sajjadi MSM, Scholkopf B, Hirsch M. EnhanceNet: single image super-resolution through automated texture synthesis. Proc IEEE Int Conf Comput Vis. (ICCV). 2017;4491–500.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 3rd Int Conf Learn Represent ICLR 2015 - Conf Track Proc. 2015;1–14.
Zaker N, Haddad K, Faghihi R, Arabi H, Zaidi H. Direct inference of Patlak parametric images in whole-body PET/ CT imaging using convolutional neural networks. Eur J Nucl Med Mol Imaging. 2022. https://doi.org/10.1007/s00259-022-05867-w.
Huang Z, Wu Y, Fu F, Meng N, Gu F, Wu Q, et al. Parametric image generation with the uEXPLORER total-body PET/CT system through deep learning. Eur J Nucl Med Mol Imaging. 2022;2482–92.
Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. MICCAI. Lect Notes Comput Sci. 2015;351. https://doi.org/10.1007/978-3-319-24574-4_28.
Cui J, Gong K, Guo N, Kim K, Liu H, Li Q. CT-guided PET parametric image reconstruction using deep neural network without prior training data. Proc Med Imag: Phys Med Imag. 2019;34.
Xie N, Gong K, Guo N, Qin Z, Wu Z, Liu H, et al. Rapid high-quality PET Patlak parametric image generation based on direct reconstruction and temporal nonlocal neural network. Neuroimage. Elsevier. 2021;240:118380.
Zhang X, Cherry SR, Xie Z, Shi H, Badawi RD, Qi J. Subsecond total-body imaging using ultrasensitive positron emission tomography. Proc Natl Acad Sci U S A. 2020;117:2265–7.
Wang Y, Yu B, Wang L, Zu C, Lalush DS, Lin W, et al. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage. 2018;174:550–62.
Funding
This work was supported in part by the National Key Research and Development Program of China (No: 2020AAA0109502), by the National Natural Science Foundation of China (No: U1809204, 61701436), Zhejiang Provincial Natural Science Foundation of China (No: LY22F010007), by the Talent Program of Zhejiang Province (2021R51004), and by the Key Research and Development Program of Zhejiang Province (No: 2021C03029). Swiss National Science Foundation (SNF No. 188914) and Germaine de Staël Programm of Swiss Academy of Engineering Science (SATW).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. H. Sari and S. Xue acquired and pre-processed the data, Y. Li trained network and analyzed data, J.Hu segmented the lesions, and R. Ma and S. Kandarpa assisted in network training. Y. Li drafted the manuscript and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
The local Institutional Review Board approved the study (KEK 2019–02193), and written informed consent was obtained from all patients. The study was performed in accordance with the Declaration of Helsinki.
Conflict of interest
H. Sari is a full-time employee of Siemens Healthineers. The other authors have no conflicts of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Hu, J., Sari, H. et al. A deep neural network for parametric image reconstruction on a large axial field-of-view PET. Eur J Nucl Med Mol Imaging 50, 701–714 (2023). https://doi.org/10.1007/s00259-022-06003-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-022-06003-4