Abstract
Purpose
Small-animal single photon emission computed tomography (SPECT) with focused multi-pinhole collimation geometries allows scanning modes in which large amounts of photons can be collected from specific volumes of interest. Here we present new tools that improve targeted imaging of specific organs and tumours, and validate the effects of improved targeting of the pinhole focus.
Methods
A SPECT system with 75 pinholes and stationary detectors was used (U-SPECT-II). An XYZ stage automatically translates the animal bed with a specific sequence in order to scan a selected volume of interest. Prior to stepping the animal through the collimator, integrated webcams acquire images of the animal. Using sliders, the user designates the desired volume to be scanned (e.g. a xenograft or specific organ) on these optical images. Optionally projections of an atlas are overlaid semiautomatically to locate specific organs. In order to assess the effects of more targeted imaging, scans of a resolution phantom and a mouse myocardial phantom, as well as in vivo mouse cardiac and tumour scans, were acquired with increased levels of targeting. Differences were evaluated in terms of count yield, hot rod visibility and contrast-to-noise ratio.
Results
By restricting focused SPECT scans to a 1.13-ml resolution phantom, count yield was increased by a factor 3.6, and visibility of small structures was significantly enhanced. At equal noise levels, the small-lesion contrast measured in the myocardial phantom was increased by 42%. Noise in in vivo images of a tumour and the mouse heart was significantly reduced.
Conclusion
Targeted pinhole SPECT improves images and can be used to shorten scan times. Scan planning with optical cameras provides an effective tool to exploit this principle without the necessity for additional X-ray CT imaging.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Molecular imaging has proven to be extremely valuable in studying animal models of human disease and in the development of new pharmaceuticals and tracers. Many molecular mechanisms can be assessed quantitatively in vivo using radionuclide techniques such as single photon emission computed tomography (SPECT) and positron emission tomography (PET). In the past, SPECT lacked the resolution necessary to accurately image organs of small animals such as mice and rats. Several newly developed dedicated small-animal SPECT systems have overcome this limitation [1–12].
Recently, sub-half-millimetre image resolution has been achieved in SPECT, using multi-pinhole collimators combined with high pinhole magnification factors [1, 3, 13–15]. As these systems are equipped with collimators that have pinholes that focus on a central area in the imaging cavity, the fraction of detected photons from specific organs or tissue of interest is very high. This can result in improved noise resolution trade-offs over systems with a lower level of focusing, and the possibility of reducing the tracer dose or the acquisition time. Using focused pinhole geometries, detailed images of mouse and rat organs (e.g. beating heart, kidney and the brain) and tumours have been acquired [1, 3, 13, 15–18].
Multi-pinhole collimators with focused geometries are also able to scan larger volumes – up to the total body of mice and rats. This is accomplished by translating the animal through the collimator in concert with specially adapted reconstruction methods that use projections from all bed positions simultaneously [3, 14]. To increase count yield from a specific organ or tumour, the field-of-view (FOV) of a system with a highly focused geometry and an XYZ stage can be confined to a region that mainly contains the tissue of interest (“sensitivity painting”). This requires making an estimate of the location of these tissues. Disadvantages to performing this estimation based on X-ray CT images include additional radiation dose, hardware, and scan time. Localization based on MRI, which is currently an area of active research, also requires additional hardware and scan time. Furthermore, using pinhole projection images combined with a persistence scope, accurate localization is difficult to achieve since there is a small FOV, few gamma photons can be detected in a limited time and the tissue being localized may have a very low uptake. The aim of the present study was to explore the alternative possibility of using low-cost optical cameras for tissue localization and FOV selection, and to empirically investigate the effects of targeting on sensitivity and the quality of reconstructed images.
Materials and methods
SPECT system with optical cameras
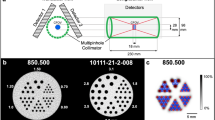
U-SPECT-II [3, 19] (Fig. 1a) is a multi-pinhole SPECT scanner for imaging rodents. It consists of three stationary detector arrays and exchangeable collimators for different sized animals or for specific organs such as the brain [20]. Available collimators consist of a tungsten cylinder containing 75 micro-pinhole apertures that together provide a FOV the shape of which is illustrated in Fig. 1c. The pinhole geometry is chosen such that the region observed through all pinholes simultaneously is located in the centre of the collimator. For this part of the FOV, referred to as the central FOV (CFOV, Fig. 1c) and a small area around it, complete data are acquired without any translation of the bed during scanning. For the mouse collimators in this study, such as the general purpose mouse collimator (MILabs, Utrecht, The Netherlands), the CFOV is approximately a cylinder of length 7 mm and diameter 12 mm. For the rat collimators these dimensions are about twice as large.
a U-SPECT-II system with close-ups of a mouse on the animal bed with a transparent heater pad in front of optical cameras and the graphical user interface with optical images and three real-time projection images. b Example of various FOV selection boundaries in three dimensions. c Schematic cross-sections of FOV and CFOV in U-SPECT-II general-purpose collimators
The FOV outside the CFOV also contributes to the projection data, but in order to correctly reconstruct volumes significantly larger than the CFOV, the system must move the focus over the region of interest. This scanning focus method [14] enables sensitivity painting similar to dose painting in radiotherapy. The bed is mounted on a motor-controlled XYZ stage, which allows accurate positioning of the animal. During SPECT acquisition, the XYZ stage automatically moves the animal stepwise through the collimator, thereby effectively moving the CFOV over the animal in order to obtain complete data for any part of the animal that is selected by the user. The bed is transparent and has a half-cylindrical shape. It contains a transparent heater pad to control the temperature of the animal (Fig. 1a).
Description of the FOV selection tool
Optical image-based positioning
Prior to acquiring SPECT data, three optical cameras, which are integrated with the U-SPECT-II system, take photographs of the animal from the left, top and right (Fig. 1a). The cameras (DFK 21F04; The Imaging Source, Germany) are equipped with a quarter-inch CCD detector with a resolution of 640 × 480 pixels. The photographs are displayed on a graphical user interface, on which the user can define a box to be scanned (Fig. 1a, b). Next, the software calculates a sequence of bed positions in such a way that the volume within the box will be sampled by the CFOV in at least one of the bed positions. The user can optionally check and fine-tune the position of the FOV using real-time gamma photon projection images of the centre of the selected FOV, obtained through pinholes that provide views of the animal at approximately the same angles as the optical cameras. Depending on the size of the selection, the number of positions can range from one or two positions (for organs such as the heart, the brain or a tumour) up to tens of positions for total-body scanning. Since changing the bed position takes only 0.7 s on average, even for total-body studies fast dynamic acquisitions are possible using the scanning focus method [15, 21].
Atlas overlay
Our optical image-based FOV selection allows targeting SPECT to a joint, a xenograft (Fig. 2c) or any other part of the animal. To further aid localization of organs, projections of an anatomical atlas that shows the different organs of an animal can be projected onto the optical images (Fig. 2d). The overlay is based on projections of a manually segmented MR image. It can be manually scaled to register it to the optical images, for example using the nose, the eyes, the shoulders, the root of the tail and the skin contour as reference features. To account for different animal shapes and positioning, the atlas projections can also be deformed in the transaxial directions by repositioning markers on the skin contour. This method provides an approximation of the organ positions, which can be very useful as a guideline during scan planning since the (rough) location of organs can easily be mistaken.
Different volume selections applied to obtain a specific SPECT acquisition of (a) a resolution phantom, (b) a mouse myocardial phantom, (c) a mouse tumour, and (d) a mouse heart using an atlas. Red (large) box nontargeted selection, orange (middle) box selection targeted in one dimension only, yellow (small) box selection targeted in three dimensions
Aligning optical images to SPECT images
The mapping of the optical images (and consequently also the FOV selection) to the SPECT reconstructed voxel grid is determined by calibration. Before calibration, the optical cameras were corrected for rotation, scaling and barrel distortions based on optical images of a millimetre grid. In order to reduce perspective errors and approximate parallel projections, the optical images were acquired as a set of small image strips that were stitched together. The calibration was performed by scanning a phantom containing several point sources and registering its optical images to maximum intensity projections of the SPECT volume. To create the point sources, ion exchange resin beads with a diameter of approximately 0.2 mm were dipped in a mixture of 99mTc-pertechnetate and ink, to make them visible in both modalities.
The optical-to-SPECT registration was performed by applying a rigid transformation that minimizes the mean distance between the optical point sources and their corresponding point sources in the maximum intensity projection of the SPECT volume. After applying this transformation to our system, the maximum distance between any of the point sources in the optical and SPECT images was 0.25 mm.
Validation
Improvements due to restricting the scan area, in terms of sensitivity and resolution, were determined with a resolution phantom, whereas improvements in contrast-to-noise ratio were measured in scans of a mouse myocardial phantom containing a cold lesion. Finally, in vivo studies illustrated the effects of scan area size in mouse tumour and myocardial perfusion imaging.
All scans discussed here were performed three times, each time employing a different FOV selection: (1) a nontargeted scan, (2) a 1D-targeted scan (with the FOV only restricted in the z-dimension), emulating a system that can only target in the axial direction, and (3) a 3D-targeted scan with the FOV restricted in the x-, y- and z-dimensions (Fig. 2).
Resolution phantom study
In order to assess the visibility of small details, a Jaszczak-style resolution phantom (ultra-high-resolution micro-phantom 850.100; VANDERWILT techniques, Boxtel, The Netherlands) with six sections containing capillaries with diameters of 0.35, 0.40, 0.45, 0.50, 0.60 and 0.75 mm was imaged. In this phantom, the distance between the capillaries in each section equals the capillary diameter in that section. The phantom was filled with 145 MBq 99mTc-pertechnetate. The resolution phantom and the selected volumes for the different protocols are shown in Fig. 2a.
A 10-min acquisition was performed for each of the protocols (ultra-high-resolution study) and a second acquisition series with the same phantom was performed with lower activity by repeating the three scans after 20 h (high-resolution study). The ultra-high-resolution and high-resolution studies were performed with, respectively, the 0.35-mm and 0.6-mm diameter pinhole mouse collimators [3]. Images were reconstructed on a 0.1875 mm isotropic voxel grid with ten iterations pixel-based ordered subset expectation maximization (POSEM) with 16 subsets [22]. Furthermore, the total number of detected photons was determined for each scan in a 20% energy window around 140 keV to estimate the sensitivity gain achieved by restricting the scan volume. The count totals were corrected for background radiation by subtracting the number of counts detected in a separate background acquisition.
Mouse myocardial phantom study
In order to quantify improvements in signal-to-noise ratio by restricting the scan volume, a physical cardiac phantom was used. This phantom, made of polymethyl methacrylate, modelled perfusion of the left ventricular myocardium of a mouse (Fig. 3a). It contained a cavity into which a polymethyl methacrylate insert mimicking an infarct could be placed. The cavity resembled the left ventricular myocardial wall, which was filled with 7.2 MBq 99mTc-pertechnetate. The dimensions of the myocardium in the phantom (Fig. 3b) represented the average end-diastolic dimensions reported by Wiesmann et al. [23], which were measured in MRI data from 15 adult C57BL/6 mice at rest. The FOV selections for the applied scan protocols are shown in Fig. 2b.
a Photograph of a mouse myocardial phantom with lesion. b Schematic drawing of a phantom showing the dimensions of the left ventricle and lesion in millimetres. c Reconstructed short-axis slice with profile range used for circumferential profiles. d Different regions used for calculation of noise and contrast (solid areas “non-infarcted” regions, dashed area “infarcted” region)
The phantom was scanned using each of the three FOV selection protocols. The duration of the first scan was 60 min, and the next scans were slightly extended to correct for decay. Each of the three list-mode datasets was split into 60 noise realizations each containing the same number of list-mode events, which were spread out regularly over the entire scan time. This emulates 3 × 60 acquisitions with a left ventricular uptake of 0.12 MBq. Because the projections obtained each contained only 1/60 of the usual amount of background counts, projections from 60 separate 60-min background acquisitions without a phantom were added to the noisy phantom projections before reconstruction. The uptake value of 0.12 MBq was the average reconstructed left ventricular uptake measured in two mouse 99mTc-tetrofosmin scans after applying attenuation correction as described previously [24]. For each targeting level, ten noise realizations were reconstructed on a 0.1875-mm isotropic voxel grid. Maximum likelihood expectation maximization instead of POSEM was used for reconstruction because the former updates the image with smaller increments, and therefore allows for constructing plots of contrast-to-noise ratios based on a higher number of stages of convergence.
In vivo animal studies
Figure 2c, d shows the three FOV selection protocols applied to, respectively, a mouse tumour scan and a myocardial perfusion scan. All procedures employed in these studies were approved by the local ethics committee and were performed in accordance with international guidelines on handling laboratory animals.
For the tumour scan, a 21-g female mouse (CB17/SCID) was used which had a 0.5–1.0 ml A431 human carcinoma on its right flank. The scans were acquired under isoflurane anaesthesia 3 days after injection of 49 MBq 111In-labelled Unibody (Genmab, The Netherlands) using the 0.6-mm diameter pinhole mouse collimator tube [3]. The duration of each acquisition was 45 min. The images were reconstructed on a 0.1875-mm isotropic voxel grid employing six iterations POSEM with 16 subsets [22]. The reconstructed images were postfiltered using a gaussian filter with σ = 0.1875 mm.
For the mouse cardiac perfusion study, a 29-g male mouse (C57BL/6J) was anaesthetized with isoflurane and injected with 134 MBq 99mTc-tetrofosmin. At 30 min after injection, the first SPECT scan of 45 min was acquired. The other two scans were corrected for decay of the isotope by adjusting the duration of each acquisition. The scans were acquired using the 0.6-mm diameter pinhole mouse collimator tube [3] and reconstructed on a 0.1875-mm isotropic voxel grid using four iterations POSEM with 16 subsets [22], employing cardiac gating with 16 intervals. The reconstructed images were postfiltered with a gaussian filter in time (σ = 1.27 time intervals) and space (σ = 0.75 mm).
Data analysis of the mouse myocardial phantom
To assess differences in contrast-to-noise ratio, small-lesion contrast and noise were measured in the reconstructed myocardial phantom images after each iteration. Contrast was then plotted as a function of noise for each targeting level. Eleven volumetric regions of interest of equal size were defined (Fig. 3d), one within the lesion and ten within the “fully perfused myocardial tissue”. The average contrast was defined as the average contrast over all noise realizations, with the contrast C of one noise realization defined as:
where M equals the number of fully perfused regions, A i is the average intensity of the lesion and A m is the average intensity of fully perfused region m. The normalized standard deviation (NSD) was used as a measure of the noise of a perfused region:
where A(k) represents the average intensity of the region in noise realization k, \( \left\langle A \right\rangle \) is the intensity of the region averaged over all noise realizations and N is the number of noise realizations. Average noise over the non-infarcted myocardium was calculated by averaging these NSD values.
Reconstructed images of the mouse myocardial phantom were postfiltered with a gaussian filter (σ = 0.1875 mm) and profiles were generated by dividing the profile range (depicted in Fig. 3c) into 36 segments and calculating the mean voxel value for each segment. In addition to plotting the profiles separately, the profiles for all ten noise realizations were also averaged into a mean profile. The variation from the mean was visualized by including two profiles denoting the mean plus and minus one standard deviation.
Results
Resolution phantom study
Figure 4 presents reconstructed image slices from the resolution phantom scans. No postfilter was applied. The displayed slice thickness is 0.75 mm for the high-resolution scans and 0.375 mm for the ultra-high-resolution scans. In both the high- and the ultra-high-resolution scans, restricting the FOV resulted in reduced noise and more sections having visually distinguishable rods. In the images obtained with the 0.6-mm pinholes and nontargeted scanning, the 0.6-mm rods were hardly visible. One-dimensional targeting made all 0.5-mm rods clearly visible. With 3D-targeting, the 0.45-mm rods seemed to be much better separated. In the ultra-high-resolution study (0.35-mm pinholes), the 0.35-mm segment could be resolved in the 3D-targeted image whereas in the 1D-targeted image it appeared less clear. In the nontargeted image, even some of the 0.45-mm rods were hardly distinguishable.
Table 1 shows the number of counts measured within a 20% energy window around 140 keV for each of the targeting levels. The number of recorded counts increased with better volume targeting as the phantom was positioned inside the CFOV more often. Since there was no 99mTc-pertechnetate outside the phantom, the increase in scan sensitivity can be estimated by comparing the total number of detected photons obtained using each of the FOV selection protocols. Compared to a nontargeted scan, 3D targeting was able to increase the number of counts detected from a specific volume of interest by a factor of approximately 3.6.
Mouse myocardial phantom study
To provide an additional measure of improvement in image quality, Fig. 5 compares contrast-to-noise ratio curves for each of the three targeting levels. For all targeting levels, average contrast improved with an increasing number of iterations, but average noise was also increased. Compared at an equal noise level of 0.076, the contrast-to-noise level achieved with 1D targeting was improved by 19% over the nontargeted reconstruction, whereas 3D targeting resulted in an improvement of 42% over the nontargeted reconstruction. Comparing images at an equal average contrast level of 0.75, 1D and 3D targeting improved the contrast-to-noise level by, respectively, 37% and 167% over the level measured in the nontargeted reconstruction. This noise reduction is illustrated in Fig. 6. The profiles in the middle row show that the variance over the ten reconstructions was reduced with more targeting, which implies improved reproducibility of intensity in individual segments. This effect is also apparent in the bottom row, which shows three examples of circular profiles. Better targeting reduced the differences between these reconstructions. In addition, the bottom row shows that better targeting resulted in less erroneous intensity variations within the myocardial wall. This is also reflected in the images in the top row, which appear much less noisy with more accurate targeting.
Average defect contrast as a function of average noise in the mouse myocardial phantom. Curves were generated by interpolating values found at different iterations (dash-dots nontargeted, dashes 1D-targeted, solid 3D-targeted). Comparisons at equal contrast and at equal noise, as described in the text, are illustrated using markers connected by dotted lines
Images and circumferential profiles of reconstructed short-axis slices of the mouse myocardial phantom for nontargeted acquisition (left), 1D-targeted acquisition (centre) and 3D-targeted acquisition (right) show stronger noise reduction with a higher degree of targeting. a Short-axis slices from reconstruction of one noise realization. b Profiles of mean (solid) and mean ±1SD (dashed), both calculated over ten noise realizations. c Example profiles from reconstructions of three different noise realizations
In vivo animal studies
Slices through the reconstructed volumes from the myocardial perfusion and tumour scans are shown in Fig. 7. Displayed slice thicknesses are 0.56 mm and 0.75 mm for, respectively, the myocardial and the tumour study. The 3D-targeted scans appeared to have less noise than the 1D-targeted and nontargeted images. This effect was most apparent in the myocardial perfusion study, where the number of counts in individual frames was low because of gating. The myocardial images from the 3D-targeted scan best revealed the structure of the papillary muscles and the right ventricular wall.
Discussion
A navigation and selection tool for SPECT acquisition based on optical imaging was developed to increase count yield from specific organs and tissues of interest. The results reported here show that targeting in three dimensions is important, because it improves count yield and contrast-to-noise trade-off.
When a large FOV is required, such as in total-body scanning, the level of targeting may become low. This results in reduced sensitivity as the total scan time is distributed over a large number of CFOV positions. Previously we and others have shown that in such situations the count rate can still be high enough to obtain excellent images [14] even for gated total-body SPECT or for studies which employ low doses [3]. Furthermore, although the overall sensitivity in total body scanning may then become similar to non-focused geometries, the combination of focusing and moving the bed in three dimensions effectively translates the axis of rotation and may therefore have the advantage of acquiring more angle information compared to detectors rotating around one longitudinal axis. In the recently launched U-SPECT-II/CT and VECTor/CT (a combined SPECT/PET/CT device with sub-millimetre resolution [25, 26]), FOV selection can alternatively be performed based on X-ray planar images, with or without the option of registering projections of an atlas to them. It should, however, be noted that these devices are also equipped with webcams for, for example, FOV selection.
Using optical cameras with the option of registering projections of an atlas to the optical images is a cost-effective method and has many advantages over scan planning using an additional CT scan. First, the limited tissue contrast in a CT scan may hamper accurate localization for many organs, whereas our atlas images are already presegmented. Furthermore, X-ray imaging exposes the animal to an extra radiation dose [27–29]. Another advantage of recording optical images is that, after correcting for differences in positioning and distortion of the lens, they can be related directly to images obtained using other optical modalities such as bioluminescence or fluorescence. Additional CT scans or scans from other anatomic modalities can be useful in particular for the localization of activity in unknown internal structures such as a tumour.
In this study we used a basic atlas containing brain, heart, lungs, liver, spleen and kidneys, based on a mouse MRI scan. In practice, the localization of the organs worked well in most scans, especially since the posture of the animal was chosen to be close to that of the mouse in the MRI scan. More extensive in vivo position verification may be an interesting subject for future studies. In addition, atlases and anatomical images based on MRI or other modalities are being created, in which the animals can be placed in many different postures. Furthermore, other atlases of mice and rats are available [30–35]. Even when using accurate atlases, small localization errors may remain as a result of individual differences or pathologies. An atlas that can be automatically deformed to match the optical image, based on thin-plate spline deformations, is currently being investigated [36, 37].
Conclusion
Focused multi-pinhole geometries combined with an XYZ translation stage give the opportunity to acquire a high number of counts from the organ of interest. As a result, much smaller pinholes can be used to obtain ultra-high-resolution SPECT images or images with reduced noise. The results of the present study show that count yield increases dramatically when targeting is applied, which results in new opportunities for fast dynamic imaging of tumours or organs. This new method could also be applied to increasing throughput or to reducing radiation doses. We have developed a fast and user-friendly tool for estimating organ positions based on optical images and optional atlases. This tool allows maximal benefit to be obtained from the unique advantages of SPECT with focused multi-pinhole collimators.
References
Beekman FJ, van der Have F, Vastenhouw B, van der Linden AJA, van Rijk PP, Burbach JPH, et al. U-SPECT-I: A novel system for submillimeter-resolution tomography with radiolabeled molecules in mice. J Nucl Med 2005;46(7):1194–200.
Furenlid LR, Wilson DW, Chen YC, Kim H, Pietraski PJ, Crawford MJ, et al. A second-generation high-resolution dynamic SPECT imager. IEEE Trans Nucl Sci 2004;51(3):631–5.
Van der Have F, Vastenhouw B, Ramakers RM, Branderhorst W, Krah JO, Ji C, et al. U-SPECT-II: an ultra-high resolution device for molecular small-animal imaging. J Nucl Med 2009;50(4):599–605.
Goertzen AL, Jones DW, Seidel J, Li K, Green MV. First results from the high-resolution mouseSPECT annular scintillation camera. IEEE Trans Med Imaging 2005;24(7):863–7.
Ishizu K, Mukai T, Yonekura Y, Pagani M, Fujita T, Magata Y, et al. Ultra-high-resolution SPECT system using four pinhole collimators for small animal studies. J Nucl Med 1995;36(12):2282–7.
Jaszczak RJ, Li J, Wang H, Zalutsky MR, Coleman RE. Pinhole collimation for ultra-high-resolution small-field-of-view SPECT. Phys Med Biol 1994;39(3):425–37.
King MA, Pretorius PH, Farncombe T, Beekman FJ. Introduction to the physics of molecular imaging with radioactive tracers in small animals. J Cell Biochem Suppl 2002;39:221–30.
McElroy DP, MacDonald LR, Beekman FJ, Wang YC, Patt BE, Iwanczyk JS, et al. Performance evaluation of A-SPECT: a high resolution desktop pinhole SPECT system for imaging small animals. IEEE Trans Nucl Sci 2002;49:2139–47.
Meikle SR, Kench PL, Weisenberger AG, Wojcik R, Smith MF, Majewski S, et al. A prototype coded aperture detector for small animal SPECT. IEEE Trans Nucl Sci 2002;49(5):2167–71.
Palmer J, Wollmer P. Pinhole emission computed tomography: method and experimental evaluation. Phys Med Biol 1990;35(3):339–50.
Schramm NU, Ebel G, Engeland U, Schurrat T, Béhé M, Behr TM. High-resolution SPECT using multipinhole collimation. IEEE Trans Nucl Sci 2003;50(3):315–20.
Wu MC, Tang HR, Gao DW, Ido A, O’Connell JW, Hasegawa BH, et al. ECG-gated pinhole SPECT in mice with millimeter resolution. IEEE Trans Nucl Sci 2000;47(3):1218–27.
Beekman F, van der Have F. The pinhole: gateway to ultra-high resolution three-dimensional radionuclide imaging. Eur J Nucl Med Mol Imaging 2007;34(2):151–61.
Vastenhouw B, Beekman F. Submillimeter total-body murine imaging with U-SPECT-I. J Nucl Med 2007;48(3):487–93.
Vastenhouw B, van der Have F, van der Linden AJA, von Oerthel L, Booij J, Burbach JPH, et al. Movies of dopamine transporter occupancy with ultra-high resolution focusing SPECT. Mol Psychiatr 2007;12:984–7.
Wyckhuys T, Staelens S, Van Nieuwenhuyse B, Deleye S, Hallez H, Vonck K, et al. Hippocampal deep brain stimulation induces decreased rCBF in the hippocampal formation of the rat. Neuroimage 2010;52(1):55–61.
De Bruyne S, Wyffels L, Boos TL, Staelens S, Deleye S, Rice KC, et al. In vivo evaluation of [123I]-4-(2-(bis(4-fluorophenyl)methoxy)ethyl)-1-(4-iodobenzyl)piperidine, an iodinated SPECT tracer for imaging the P-gp transporter. Nucl Med Biol 2010;37(4):469–77.
Van Steenkiste C, Staelens S, Deleye S, De Vos F, Vandenberghe S, Geerts A, et al. Measurement of porto-systemic shunting in mice by novel three-dimensional micro-single photon emission computed tomography imaging enabling longitudinal follow-up. Liver Int 2010;30(8):1211–20.
Beekman FJ. Radiation detection device, scintillation device and detection method, as well as multiple image-forming device. PCT/NL2006/000513. April 19, 2007.
Beekman FJ, Vastenhouw B, van der Wilt G, Vervloet M, Visscher R, Booij J, et al. 3-D rat brain phantom for high-resolution molecular imaging. Proc IEEE 2009;97(12):1997–2005.
Vastenhouw B, Ramakers R, Beekman F. High resolution dynamic total-body animal imaging with U-SPECT-II. J Nucl Med 2009;50(Suppl 2):526.
Branderhorst W, Vastenhouw B, Beekman FJ. Pixel-based subsets for rapid multi-pinhole SPECT reconstruction. Phys Med Biol 2010;55(7):2023–34.
Wiesmann F, Ruff J, Engelhardt S, Hein L, Dienesch C, Leupold A, et al. Dobutamine-stress magnetic resonance microimaging in mice: acute changes of cardiac geometry and function in normal and failing murine hearts. Circ Res 2001;88(6):563–9.
Wu C, Van der Have F, Vastenhouw B, Dierckx RAJO, Paans AMJ, Beekman FJ. Absolute quantitative total-body small-animal SPECT with focusing pinholes. Eur J Nucl Med Mol Imaging 2010;37(11):2127–35.
Goorden MC, Beekman FJ. High-resolution tomography of positron emitters with clustered pinhole SPECT. Phys Med Biol 2010;55(5):1265–77.
Beekman FJ, van der Have F, Kreuger R, Goorden MC. Simultaneous sub-millimetre PET and SPECT with a dedicated multi-pinhole geometry. 2010 World Molecular Imaging Congress. Kyoto, Japan.
Carlson S, Classic K, Bender C, Russell S. Small animal absorbed radiation dose from serial micro-computed tomography imaging. Mol Imaging Biol 2007;9(2):78–82.
Boone JM, Velazquez O, Cherry SR. Small-animal X-ray dose from micro-CT. Mol Imaging 2004;3(3):149–58.
Figueroa SD, Winkelmann CT, Miller HW, Volkert WA, Hoffman TJ. TLD assessment of mouse dosimetry during microCT imaging. Med Phys 2008;35(9):3866–74.
Segars WP, Tsui BMW, Frey EC, Johnson GA, Berr SS. Development of a 4-D digital mouse phantom for molecular imaging research. Mol Imaging Biol 2004;6(3):149–59.
Dogdas B, Stout D, Chatziioannou AF, Leahy RM. Digimouse: a 3D whole body mouse atlas from CT and cryosection data. Phys Med Biol 2007;52(3):577–87.
Stabin MG, Peterson TE, Holburn GE, Emmons MA. Voxel-based mouse and rat models for internal dose calculations. J Nucl Med 2006;47(4):655–9.
Bai X, Yu L, Liu Q, Zhang J, Li A, Han D, et al. A high-resolution anatomical rat atlas. J Anat 2006;209(5):707–8.
Wu L, Zhang G, Luo Q, Liu Q. An image-based rat model for Monte Carlo organ dose calculations. Med Phys 2008;35(8):3759–64.
Keenan MA, Stabin MG, Segars WP, Fernald MJ. RADAR realistic animal model series for dose assessment. J Nucl Med 2010;51(3):471–6.
Baiker M, Dijkstra J, Que I, Löwik CWGM, Reiber JHC, Lelieveldt BPF. Organ approximation in micro-CT data with low soft tissue contrast using an articulated whole-body atlas. Proceedings of the 5th IEEE International Symposium on Biomedical Imaging. Paris, France. 2008;1267–70.
Baiker M, Vastenhouw B, Branderhorst W, Reiber JHC, Beekman F, Lelieveldt BPF. Atlas-driven scan planning for high-resolution μSPECT scanning from multi-view photographs: a pilot study. Proceedings of SPIE Medical Imaging 2009. Lake Buena Vista (Orlando), Florida. 2009; paper #72611L, 8 pages.
Acknowledgments
We thank Ruud Ramakers (UMC Utrecht) and Jeroen van den Brakel (Genmab) for their technical support and help with measurements, and Jan van Ewijk, Erwin Bakker and Jesse Bosma (UMC Utrecht) for their help in designing and manufacturing the mouse myocardial phantom.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Branderhorst, W., Vastenhouw, B., van der Have, F. et al. Targeted multi-pinhole SPECT. Eur J Nucl Med Mol Imaging 38, 552–561 (2011). https://doi.org/10.1007/s00259-010-1637-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-010-1637-4