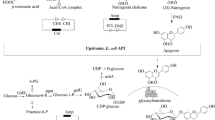
Abstract
Flavonoids are a large family of plant and fungal natural products, among which many have been found to possess outstanding biological activities. Utilization of engineered microbes as surrogate hosts for heterologous biosynthesis of flavonoids has been investigated extensively. However, current microbial biosynthesis strategies mostly rely on using one microbial strain to accommodate the long and complicated flavonoid pathways, which presents a major challenge for production optimization. Here, we adapt the emerging modular co-culture engineering approach to rationally design, establish and optimize an Escherichia coli co-culture for de novo biosynthesis of flavonoid sakuranetin from simple carbon substrate glucose. Specifically, two E. coli strains were employed to accommodate the sakuranetin biosynthesis pathway. The upstream strain was engineered for pathway intermediate p-coumaric acid production, whereas the downstream strain converted p-coumaric acid to sakuranetin. Through step-wise optimization of the co-culture system, we were able to produce 29.7 mg/L sakuranetin from 5 g/L glucose within 48 h, which is significantly higher than the production by the conventional monoculture-based approach. The co-culture biosynthesis was successfully scaled up in a fed-batch bioreactor, resulting in the production of 79.0 mg/L sakuranetin. To our knowledge, this is the highest bioproduction concentration reported so far for de novo sakuranetin biosynthesis using the heterologous host E. coli. The findings of this work expand the applicability of modular co-culture engineering for addressing the challenges associated with heterologous biosynthesis of complex natural products.
Key points
• De novo biosynthesis of sakuranetin was achieved using E. coli-E. coli co-cultures.
• Sakuranetin production by co-cultures was significantly higher than the mono-culture controls.
• The co-culture system was optimized by multiple metabolic engineering strategies.
• The co-culture biosynthesis was scaled up in fed-batch bioreactor.





Similar content being viewed by others
References
Camacho-Zaragoza JM, Hernandez-Chavez G, Moreno-Avitia F, Ramírez-Iñiguez R, Martínez A, Bolívar F, Gosset G (2016) Engineering of a microbial coculture of Escherichia coli strains for the biosynthesis of resveratrol. Microb Cell Factories 15(1):163. https://doi.org/10.1186/s12934-016-0562-z
Chen T, Zhou Y, Lu Y, Zhang H (2019) Advances in heterologous biosynthesis of plant and fungal natural products by modular co-culture engineering. Biotechnol Lett 41(1):27–34. https://doi.org/10.1007/s10529-018-2619-z
Davis JH, Rubin AJ, Sauer RT (2010) Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res 39(3):1131–1141. https://doi.org/10.1093/nar/gkq810
Ganesan V, Li Z, Wang X, Zhang H (2017) Heterologous biosynthesis of natural product naringenin by co-culture engineering. Synth Syst Biotechnol 2(3):236–242. https://doi.org/10.1016/j.synbio.2017.08.003
Gasser B, Saloheimo M, Rinas U, Dragosits M, Rodríguez-Carmona E, Baumann K, Giuliani M, Parrilli E, Branduardi P, Lang C (2008) Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Factories 7(1):11. https://doi.org/10.1186/1475-2859-7-11
Grecco SS, Reimão JQ, Tempone AG, Sartorelli P, Cunha RL, Romoff P, Ferreira MJ, Fávero OA, Lago JHG (2012) In vitro antileishmanial and antitrypanosomal activities of flavanones from Baccharis retusa DC.(Asteraceae). Exp Parasitol 130(2):141–145. https://doi.org/10.1016/j.exppara.2011.11.002
Guo X, Li Z, Wang X, Wang J, Chala J, Lu Y, Zhang H (2019) De novo phenol bioproduction from glucose using biosensor-assisted microbial co-culture engineering. Biotechnol Bioeng 116(12):3349–3359. https://doi.org/10.1002/bit.27168
Guo X, Wang X, Chen T, Lu Y, Zhang H (2020) Comparing E. coli mono-cultures and co-cultures for biosynthesis of protocatechuic acid and hydroquinone. Biochem Eng J 156:107518. https://doi.org/10.1016/j.bej.2020.107518
Jawed K, Yazdani SS, Koffas MA (2019) Advances in the development and application of microbial consortia for metabolic engineering. Metab Eng Commun:e00095. https://doi.org/10.1016/j.mec.2019.e00095
Jones JA, Wang X (2018) Use of bacterial co-cultures for the efficient production of chemicals. Curr Opin Biotechnol 53:33–38. https://doi.org/10.1016/j.copbio.2017.11.012
Jones JA, Vernacchio VR, Sinkoe AL, Collins SM, Ibrahim MH, Lachance DM, Hahn J, Koffas MA (2016) Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab Eng 35:55–63. https://doi.org/10.1016/j.ymben.2016.01.006
Jones JA, Vernacchio VR, Collins SM, Shirke AN, Xiu Y, Englaender JA, Cress BF, McCutcheon CC, Linhardt RJ, Gross RA (2017) Complete biosynthesis of anthocyanins using E. coli polycultures. MBio 8(3):e00621–e00617. https://doi.org/10.1128/mBio.00621-17
Kim M-J, Kim B-G, Ahn J-H (2013) Biosynthesis of bioactive O-methylated flavonoids in Escherichia coli. Appl Microbiol Biotechnol 97(16):7195–7204. https://doi.org/10.1007/s00253-013-5020-9
Leonard E, Lim K-H, Saw P-N, Koffas MA (2007) Engineering central metabolic pathways for high-level flavonoid production in Escherichia coli. Appl Environ Microbiol 73(12):3877–3886. https://doi.org/10.1128/AEM.00200-07
Leonard E, Yan Y, Fowler ZL, Li Z, Lim C-G, Lim K-H, Koffas MA (2008) Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol Pharm 5(2):257–265. https://doi.org/10.1021/mp7001472
Li Z, Wang X, Zhang H (2019) Balancing the non-linear rosmarinic acid biosynthetic pathway by modular co-culture engineering. Metab Eng 54:1–11. https://doi.org/10.1016/j.ymben.2019.03.002
Lim CG, Fowler ZL, Hueller T, Schaffer S, Koffas MA (2011) High-yield resveratrol production in engineered Escherichia coli. Appl Environ Microbiol 77(10):3451–3460. https://doi.org/10.1128/AEM.02186-10
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428. https://doi.org/10.1021/ac60147a030
Miyahisa I, Kaneko M, Funa N, Kawasaki H, Kojima H, Ohnishi Y, Horinouchi S (2005) Efficient production of (2S)-flavanones by Escherichia coli containing an artificial biosynthetic gene cluster. Appl Microbiol Biotechnol 68(4):498–504. https://doi.org/10.1007/s00253-005-1916-3
Miyazawa M, Kinoshita H, Okuno Y (2003) Antimutagenic activity of sakuranetin from Prunus jamasakura. J Food Sci 68(1):52–56. https://doi.org/10.1111/j.1365-2621.2003.tb14113.x
Pandey RP, Parajuli P, Koffas MA, Sohng JK (2016) Microbial production of natural and non-natural flavonoids: pathway engineering, directed evolution and systems/synthetic biology. Biotechnol Adv 34(5):634–662. https://doi.org/10.1016/j.biotechadv.2016.02.012
Roell GW, Zha J, Carr RR, Koffas MA, Fong SS, Tang YJ (2019) Engineering microbial consortia by division of labor. Microb Cell Factories 18(1):35. https://doi.org/10.1186/s12934-019-1083-3
Santos CNS (2010) Combinatorial search strategies for the metabolic engineering of microorganisms. Massachusetts Institute of Technology, Cambridge http://hdl.handle.net/1721.1/59885
Santos CNS, Koffas M, Stephanopoulos G (2011) Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab Eng 13(4):392–400. https://doi.org/10.1016/j.ymben.2011.02.002
Santos CNS, Xiao W, Stephanopoulos G (2012) Rational, combinatorial, and genomic approaches for engineering L-tyrosine production in Escherichia coli. Proc Natl Acad Sci U S A 109(34):13538–13543. https://doi.org/10.1073/pnas.1206346109
Sgobba E, Stumpf AK, Vortmann M, Jagmann N, Krehenbrink M, Dirks-Hofmeister ME, Moerschbacher B, Philipp B, Wendisch VF (2018) Synthetic Escherichia coli-Corynebacterium glutamicum consortia for l-lysine production from starch and sucrose. Bioresour Technol 260:302–310. https://doi.org/10.1016/j.biortech.2018.03.113
Shah FLA, Ramzi AB, Baharum SN, Noor NM, Goh H-H, Leow TC, Oslan SN, Sabri S (2019) Recent advancement of engineering microbial hosts for the biotechnological production of flavonoids. Mol Biol Rep 46:6647–6659. https://doi.org/10.1039/C6NP00017G
Shimizu T, Lin F, Hasegawa M, Okada K, Nojiri H, Yamane H (2012) Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice. J Biol Chem 287(23):19315–19325. https://doi.org/10.1074/jbc.M112.351270
Thuan NH, Chaudhary AK, Van Cuong D, Cuong NX (2018a) Engineering co-culture system for production of apigetrin in Escherichia coli. J Ind Microbiol Biotechnol 45(3):175–185. https://doi.org/10.1007/s10295-018-2012-x
Thuan NH, Trung NT, Cuong NX, Van Cuong D, Van Quyen D, Malla S (2018b) Escherichia coli modular coculture system for resveratrol glucosides production. World J Microbiol Biotechnol 34(6):75. https://doi.org/10.1007/s11274-018-2458-z
Wang X, Policarpio L, Prajapati D, Li Z, Zhang H (2019) Developing E. coli-E. coli co-cultures to overcome barriers of heterologous tryptamine biosynthesis. Metab Eng Commun:e00110. https://doi.org/10.1016/j.mec.2019.e00110
Wu G, Yan Q, Jones JA, Tang YJ, Fong SS, Koffas MA (2016) Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol 34(8):652–664. https://doi.org/10.1016/j.tibtech.2016.02.010
Zhang H, Stephanopoulos G (2013) Engineering E. coli for caffeic acid biosynthesis from renewable sugars. Appl Microbiol Biotechnol 97(8):3333–3341. https://doi.org/10.1007/s00253-012-4544-8
Zhang H, Stephanopoulos G (2016) Co-culture engineering for microbial biosynthesis of 3-amino-benzoic acid in Escherichia coli. Biotechnol J 11(7):981–987. https://doi.org/10.1002/biot.201600013
Zhang H, Wang X (2016) Modular co-culture engineering, a new approach for metabolic engineering. Metab Eng 37:114–121. https://doi.org/10.1016/j.ymben.2016.05.007
Zhang X, Hung TM, Phuong PT, Ngoc TM, Min B-S, Song K-S, Seong YH, Bae K (2006) Anti-inflammatory activity of flavonoids from Populus davidiana. Arch Pharm Res 29(12):1102–1108. https://doi.org/10.1007/BF02969299
Zhang L, Kong Y, Wu D, Zhang H, Wu J, Chen J, Ding J, Hu L, Jiang H, Shen X (2008) Three flavonoids targeting the β-hydroxyacyl-acyl carrier protein dehydratase from Helicobacter pylori: crystal structure characterization with enzymatic inhibition assay. Protein Sci 17(11):1971–1978. https://doi.org/10.1110/ps.036186.108
Zhang H, Pereira B, Li Z, Stephanopoulos G (2015) Engineering Escherichia coli coculture systems for the production of biochemical products. Proc Natl Acad Sci U S A 112(27):8266–8271. https://doi.org/10.1073/pnas.1506781112
Zhou K, Qiao K, Edgar S, Stephanopoulos G (2015) Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol 33(4):377–383. https://doi.org/10.1038/nbt.3095
Funding
This material is based upon work supported in part by the National Science Foundation under Grant No. 1706058. The authors received support from the startup research fund provided by Rutgers, The State University of New Jersey. ZL is a recipient of the CSC Ph.D. fellowship.
Author information
Authors and Affiliations
Contributions
XW and HZ conceived and designed research. XW, ZL, and LP conducted experiments. MK conceived research and contributed experiment resources. XW and HZ analyzed data. ZL and XW wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 250 kb)
Rights and permissions
About this article
Cite this article
Wang, X., Li, Z., Policarpio, L. et al. De novo biosynthesis of complex natural product sakuranetin using modular co-culture engineering. Appl Microbiol Biotechnol 104, 4849–4861 (2020). https://doi.org/10.1007/s00253-020-10576-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10576-1