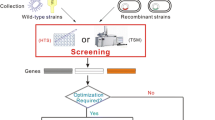
Abstract
Nitrilases are industrially important biocatalysts due to their ability to degrade nitriles to carboxylic acids and ammonia. In this study, a workflow for simple and fast recovery of nitrilase candidates from metagenomes is presented. For identification of active enzymes, a NADH-coupled high-throughput assay was established. Purification of enzymes could be omitted as the assay is based on crude extract containing the expressed putative nitrilases. In addition, long incubation times were avoided by combining nitrile and NADH conversion in a single reaction. This allowed the direct measurement of nitrile degradation and provided not only insights into substrate spectrum and specificity but also in degradation efficiency. The novel assay was used for investigation of candidate nitrilase-encoding genes. Seventy putative nitrilase-encoding gene and the corresponding deduced protein sequences identified during sequence-based screens of metagenomes derived from nitrile-treated microbial communities were analyzed. Subsequently, the assay was applied to 13 selected candidate genes and proteins. Six of the generated corresponding Escherichia coli clones produced nitrilases that showed activity and one unusual nitrilase was purified and analyzed. The activity of the novel arylacetonitrilase Nit09 exhibited a broad pH range and a high long-term stability. The enzyme showed high activity for arylacetonitriles with a KM of 1.29 mM and a Vmax of 13.85 U/mg protein for phenylacetonitrile. In conclusion, we provided a setup for simple and rapid analysis of putative nitrilase-encoding genes from sequence to function. The suitability was demonstrated by identification, isolation, and characterization of the arylacetonitrilase.
Key points
• A simple and fast high-throughput nitrilase screening was developed.
• A set of putative nitrilases was successfully screened with the assay.
• A novel arylacetonitrilase was identified, purified, and characterized in detail.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitriles are organic compounds that harbor −C≡N as functional group. Many of these compounds are toxic. They are widespread in nature and present in plants as cyanoglycosides (Conn 1979), cyanohydrins in fungi and arthropods, different antibiotics in bacteria, or ricinine and phenylacetonitrile in plants (Jallageas et al. 1980).
For the enzymatic degradation of nitriles, two pathways are known (Supplementary Fig. S1). The first one is the direct conversion of nitriles to corresponding carboxylic acids and ammonia via nitrilases (EC 3.5.5.-). The second one involves the degradation of nitriles to corresponding amides via nitrile hydratases (NHases) (EC 4.2.1.84) and the subsequent hydrolysis of amides to carboxylic acids and ammonia using amidases (EC 3.5.1.4). Respective enzymes are present in bacteria (Egelkamp et al. 2017), filamentous fungi (Martínková et al. 2009), yeasts (Rustler and Stolz 2007), and plants (Piotrowski 2008). The enzymes are used in industry for the production of bulk chemicals such as nicotinic acid or glycolic acid (Shaw et al. 2003; Panova et al. 2007). Novel nitrile-degrading enzymes, especially those with reduced substrate and/or product inhibition under production conditions, are of industrial relevance (Bui et al. 1984; Vaughan et al. 1989; Almatawah and Cowan 1999; Zhang et al. 2011a).
Many nitrile-degrading enzymes were found via (meta)genome mining techniques. A functional screening approach was applied to identify nitrilase-encoding genes from plasmid libraries containing metagenomic DNA from different sources (Robertson et al. 2004; Bayer et al. 2011; Soares Bragança et al. 2017). In addition, in silico screening of (meta)genomic sequence data was used to identify putative nitrilases and nitrile hydratases in publicly available databases (Vergne-Vaxelaire et al. 2013). Although sequence-based approaches lead very quickly to many new gene candidates for the targeted enzyme type, they remain as predictions until functional verification. To overcome this limitation and take advantage of the constantly growing metagenomic sequence pool, procedures are needed for efficient verification of nitrilase activity and identification of potential substrates of these enzymes.
The focus of this study was to establish a workflow that enables the fast functional verification of sequence-based, screening-derived putative nitrilases and their substrates. Six metagenomes, which were obtained during a study targeting the impact of different nitriles on microbial communities (Egelkamp et al. 2019), were chosen as starting material. A procedure was established to reduce the 70 putative nitrilase-encoding genes that had been inferred from sequence-driven mining of the metagenomes to candidates with verified enzyme activity. Serving as proof of concept, one enzyme candidate was heterologously expressed and purified. Subsequently, the recovered enzyme and its activity were characterized, including the determination of optimal pH, temperature range, stability, substrate specificity, and enzyme kinetics. In addition, influences of divalent ions and other substances on enzyme activity were determined.
Material and methods
Origin of nitrilases
Putative nitrilase-encoding genes were identified in, and amplified from, metagenomic DNA that originated from compost-derived enrichment cultures containing either phenylacetonitrile, succinonitrile, acetonitrile, crotononitrile, 4-hydroxybenzonitrile, or cyclohexanecarbonitrile (Egelkamp et al. 2019). The compost sample was collected in the Experimental Botanical Garden of Georg-August-Universität Göttingen (Germany; 51° 33′ 22.6″ N, 9° 57′ 16.2″ E). GenBank accession numbers of the annotated corresponding 6 metagenomes are as follows: phenylacetonitrile, RCUE00000000; succinonitrile, RCUP00000000; acetonitrile, RCUQ00000000, crotononitrile, RCUN00000000; 4-hydroxybenzonitrile, RCUF00000000, and cyclohexanecarbonitrile, RCUO00000000.
An already characterized aliphatic nitrilase from Rhodococcus rhodochrous K22 was used as positive control for establishing the high-throughput nitrilase assay (Kobayashi et al. 1990; Kobayashi et al. 1992b). For this purpose, the corresponding nitrilase gene was codon-optimized for Escherichia coli K12 derivates using the web suite JCat (http://www.jcat.de) (Grote et al. 2005) and synthesized by Integrated DNA Technologies (Leuven, Belgium).
Bioinformatic analysis of described and putative nitrilases
Nitrilase sequence reference data were obtained from the SWISS-Prot database (Bairoch and Apweiler 2000; date of search: October 14, 2018). The data were further processed by removing misannotated enzymes, mere subunits, or nitrile hydratases designated as nitrilases. Clustering of data was done with CD-HIT (Huang et al. 2010). A fasta file containing the SWISS-Prot nitrilases and metagenome-deduced nitrilases was used as input and a sequence identity cutoff of 40% was set for clustering (Supplementary Data File S1).
Growth medium
Lysogeny broth (LB) (10 g tryptone, 10 g NaCl, and 5 g yeast extract per liter) was used for growth of microorganisms. For solid media, 15 g agar per liter was added.
Amplification of metagenome-encoded putative nitrilases
PCR reaction mixture (total volume 50 μL) contained 10 μL 5-fold Phusion HF buffer, 200 μM of each dNTP, 0.2 μM of each primer (Supplementary Table S1), 3% DMSO, 50 ng metagenomic DNA as template and 1 U of Phusion polymerase (Thermo Fisher Scientific, Waltham, MA, USA). Initial denaturation was performed at 98 °C for 5 min, followed by 30 cycles of denaturation at 98 °C for 30 s, annealing (temperature based on primer melting temperature) for 30 s and elongation at 72 °C for 45 s per kbp. The final elongation was for 5 min at 72 °C.
Plasmids, strains, and transformation
Amplified putative nitrilase-encoding genes (Supplementary Data File S2) were cloned into the pBAD18 vector system (Guzman et al. 1995). Chemically competent E. coli TOP10 cells were transformed according to the protocol of the manufacturer (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, cells were plated on 100 μg/mL ampicillin-containing LB plates and incubated overnight at 37 °C. The fidelity of the constructs was checked by Sanger sequencing (Microsynth Seqlab, Göttingen, Germany).
Heterologous expression of putative nitrilases and purification of His6-tagged proteins
E. coli TOP10 strains containing the pBAD18-based recombinant plasmids were grown in LB medium at 180 rpm (Innova 44 shaker, New Brunswick Scientific, Nürtingen, Germany) and 37 °C overnight. The preculture was used to inoculate 10 mL LB medium with an OD600 of 0.1. Subsequently, the culture was incubated for 1.5 h at 37 °C and 180 rpm (Innova 44 shaker) to a final OD600 of 0.6–0.8. For induction of heterologous gene expression, 1.5% l-(+)-arabinose was added, followed by 6 h of incubation.
Cells containing the produced (His6-tagged) nitrilase were washed twice with 1× LEW buffer of the Protino Ni-TED kit (Macherey-Nagel, Düren, Germany) and resuspended in 1.5 mL of the same buffer containing 40 μg/mL DNase I and 0.1% (w/v) lysozyme. The cells were then disrupted by at least three passages through a French press at 1.38 × 108 Pa (Thermo Fisher Scientific). The extract was cleared by centrifugation at 6000×g and 4 °C for 20 min. The recovered supernatant was loaded onto Protino Ni-TED columns according to the protocol of the manufacturer (Macherey-Nagel). The purified enzyme was further analyzed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli 1970). Protein concentration was determined using the Bradford method (Bradford 1976) with bovine serum albumin as standard.
Substrates for nitrilase screenings
Stock solutions of nitriles (final concentration of 2.7 M) were generated by solving the nitriles in N,N-dimethylformamide (DMF) (Merck KGaA, Darmstadt, Germany). These stock solutions were sterile-filtered and stored at 4 °C. The following nitriles were used: phenylacetonitrile, acetonitrile (both TCI Deutschland GmbH, Eschborn, Germany), succinonitrile, crotonitrile, 4-hydroxybenzonitrile, acetone cyanohydrin, cyclohexanecarbonitrile, fumaronitrile, and 2-phenylpropionitrile (all Sigma-Aldrich Chemie GmbH, Munich, Germany). In addition, mandelonitrile, 2-phenylbutyronitrile, benzonitrile, 3-indoleacetonitrile, 2-thiopheneacetonitrile, 3-thiopheneacetonitrile, (2-chlorophenyl)acetonitrile, (3-chlorophenyl)acetonitrile, (4-chlorophenyl)acetonitrile, cinnamonitrile, 1,4-phenylenediacetonitrile, 2-naphtylacetonitrile (all Sigma-Aldrich), and 3-phenylpropiontrile (Alfa Aesar, Haverhill, MA, USA) were used for further characterization of the purified arylacetonitrilase.
High-throughput nitrilase activity assay
Degradation of nitriles was measured by monitoring the release of ammonia in a coupled enzymatic reaction (Reisinger et al. 2006). The ammonia reacts with NADH and α-ketoglutarate in the presence of a glutamate dehydrogenase from bovine liver type II (GDH; Sigma-Aldrich) to glutamate. The reduced cofactor is monitored by its absorbance change at 340 nm. For screening of putative nitrilases, the high-throughput assay of Vergne-Vaxelaire (Vergne-Vaxelaire et al. 2013) was adjusted to allow real-time measurements without time-consuming incubation steps. The assay was performed in a flat-based 96-well plate. The reaction mixture (final volume 250 μL) contained 0.5 mM NADH, 1 mM α-ketoglutarate, 37 μL of a 2.7-M nitrile stock solution (solved in DMF), 20 μg crude extract (added at the end), 1 U/mL GDH, and 50 mM Tris-HCl (pH 8.0) buffer. Crude extract containing the pBAD18 cloning vector without insert was used as negative control. The reaction mixture was incubated at 37 °C in a Synergy 2 microplate reader (BioTek Instruments GmbH, Bad Friedrichshall, Germany). The absorbance was constantly measured for 1.5 h at 340 nm. For calculation of enzyme activity, the values of the pBAD18 crude extract (negative control) were subtracted from that of the crude extract containing putative nitrilases. The activity of the nitrilase from R. rhodochrous K22 (positive control) with different substrates was described previously (Kobayashi et al. 1990; Kobayashi et al. 1992b). During the first 15 min of measurement using the control enzyme, the degradation of succinonitrile, fumaronitrile, and crotononitrile was observed and the absorbance decrease was at least 0.75. Thus, only reactions completed during the first 15 min of measurement and showing at least an absorbance decrease of 0.75 were considered as nitrilase activity and selected for the subsequent analyses.
Construction of a phylogenetic tree
For calculation of a phylogenetic nitrilase tree, MEGA X version 10.1.7 (Kumar et al. 2018) was used. Fifteen characterized nitrilases and 3 amidases were recovered from the NCBI database and used as references. Arylacetonitrilases were derived from Alcaligenes faecalis JM3 (D13419; Kobayashi et al. 1993), Pseudomonas putida MTCC 5110 (EF467660; Banerjee et al. 2009), and Pseudomonas fluorescens EBC191 (AY885240; Kiziak et al. 2005). For aliphatic nitrilases, the enzymes from Rhodococcus rhodochrous K22 (D12583; Kobayashi et al. 1992b), Comamonas testosteroni (L32589; Lévy-Schil et al. 1995), and Synechocystis sp. PCC6803 (BA000022; Heinemann et al. 2003) were used as references. Sequences for aromatic nitrilases were obtained from Rhodococcus rhodochrous J1 (D11425; Kobayashi et al. 1992a), R. rhodochrous NCIMB 11216 (AX235749; Ress-Löschke et al. 2001), and Aeribacillus pallidus RAPc8 (DQ826045; Williamson et al. 2010). References for cyanide dihydratases were taken from Pseudomonas stutzeri AK61 (KM459551; Crum et al. 2015), Bacillus pumilus C1 (AF492815; Jandhyala et al. 2003), and B. pumilus 8A3 (AF492814; Jandhyala et al. 2003). β-Cyano-l-alanine nitrilases originated from Pseudomonas protegens Pf-5 (CP000076; Paulsen et al. 2005; Howden et al. 2009), P. fluorescens Pf0-1 (CP000094; Howden et al. 2009; Silby et al. 2009), and Pseudomonas pseudoalcaligenes CECT 5344 (HG916826; Wibberg et al. 2014; Acera et al. 2017). In addition, amidases from R. rhodochrous M8 (AY971668; Riabchenko et al. 2006), P. aeruginosa PAO1 (AE004091; Ambler et al. 1987), and P. fluorescens SBW25 (AM181176; Howden et al. 2009; Silby et al. 2009) were included. The reference sequences and 6 putative nitrilases identified in this study were aligned with MUSCLE (Edgar 2004), leading to 1401 positions in the final dataset. Subsequently, evolutionary analysis was performed using the Maximum Likelihood method with the General Time Reversible model (Nei and Kumar 2000) and a “very weak” branch swap filter. Evolutionary rate differences among sites were modeled with a discrete Gamma distribution (5 categories; +G, parameter = 2.0817). The tree with the highest log likelihood (− 20,729.17) is shown. Test of phylogeny was performed by calculation of 1000 bootstrap replicates, and nodes with values below 50% were condensed for the final image.
Nitrilase characterization
The activity of a novel arylacetonitrilase (Nit09) was characterized by using a colorimetric assay for detection of ammonia (Fawcett and Scott 1960). As the reagents are temperature-sensitive, all experiments were performed in an air-conditioned laboratory at 23 °C. Nitrilase reaction was performed in a 500-μL mixture containing 25 μL nitrile (100 mM solved in DMF), 1.25 μg purified His6-tagged nitrilase, and 0.1 M citrate-phosphate buffer (pH 6.0) for 2 min at the temperature optimum of 50 °C. Then, 111 μL of this reaction was transferred into 222 μL sodium phenoxide (0.266 M phenol and 4 M NaOH), followed by 333 μL of 0.01% sodium nitroprusside and 333 μL of 0.02 N hypochlorite. Ammonia reacts with sodium phenoxide and hypochlorite to produce indophenol blue. Sodium nitroprusside improves the intensity, reproducibility, and stability of the blue color (Kaplan et al. 1965). The reaction mixture was incubated for 15 min at 27 °C in darkness. Subsequently, absorption was measured at 630 nm. To determine the substrate spectrum of Nit09, 20 mM of the tested nitrile was used and the reaction mixture was incubated for 15 min.
Nitrilase inhibitors and enzyme stability
Effects of different potential inhibitors (EDTA, DTT, H2O2, HgCl2, AgNO3, CaCl2, MnSO4, MgSO4, FeSO4, CuSO4, CoSO4, ZnCl2, NaN3, and SDS) and solvents (methanol, ethanol, glycerol, isopropanol, DMSO, acetone, chloroform, and toluene) on Nit09 activity were tested. Incubation was carried out at 50 °C for 2 min in the standard reaction mixture containing a putative inhibitor at 1 mM or 5% (v/v) and 20% (v/v) of tested organic solvent. For stability analysis, the His6-tagged purified Nit09 was stored in Protino® Ni-TED elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0) containing 1 mM NaN3 at 4 °C in the dark for 91 days. Enzymatic activity was determined under standard reaction conditions.
Determination of enzymatic constants
The steady-state kinetics for Nit09 activity was measured in the presence of GDH to detect the concomitant oxidation of NADH at 340 nm and 37 °C using a UV/Vis spectrophotometer (Cary 100 UV-Vis, Varian Medical Systems, Darmstadt, Germany). The reaction mixture (final volume 1 mL) contained 0.5 mM NADH, 1 mM α-ketoglutarate, 0.1 mM to 100 mM of phenylacetonitrile (final volume 37 μL), 5 μg purified nitrilase Nit09, 1 U/mL GDH from bovine liver type II (Sigma-Aldrich), and 0.1 M HEPES buffer (pH 8.0). Negative controls were prepared without nitrilase. To determine the turnover number and the maximum number of chemical conversions of phenylacetonitrile molecules per second that a single catalytic site will execute for a given enzyme concentration, the slope (∆A340) had to be first determined. This was done by the software of Cary WinUV version 3.0 (Varian Medical Systems). With the slope, the kcat values were determined and plotted against phenylacetonitrile concentrations. Further, the constants kcat and KM were analyzed using the Michaelis-Menten equation of the R package “drc” (Ritz et al. 2015).
Results
Optimization of the NADH-coupled nitrilase assay
Most current high-throughput nitrilase assays have drawbacks such as low sensitivity (Yazbeck et al. 2006), the requirement of specific hardware like fluorometers (Banerjee et al. 2003), or suitability only for specific nitriles (Zhu et al. 2007b). A promising approach is a NADH-coupled assay in which ammonia released by the nitrilase reaction is consumed by a glutamate dehydrogenase (GDH) in the course of the conversion of NADH to NAD. The conversion of this cofactor is monitored at 340 nm and reveals activity of the tested (putative) nitrilases (Vergne-Vaxelaire et al. 2013). However, it requires extended incubation times for nitrilase reaction and provides only qualitative information and no insights into enzymatic efficiency. We modified the assay to allow fast and simple screenings of nitrilase activity with different substrates without additional incubation steps in a single reaction mixture. Therefore, concentrations of all test ingredients were optimized to facilitate continuous measurement of NADH conversion in a 96-well microtiter plate providing first insights into the efficiency of nitrile degradation. In comparison to the method of Vergne-Vaxelaire (Vergne-Vaxelaire et al. 2013), the amount of NADH was increased to 0.5 mM for a larger measurement range and the concentration of α-ketoglutarate was reduced to prevent inhibition of the GDH reaction. These modifications were tested using crude extract containing an already characterized nitrilase from Rhodococcus rhodochrous K22 (Kobayashi et al. 1990; Kobayashi et al. 1992b) as proof of principle (Fig. 1).
Nitrile degradation by the R. rhodochrous K22 nitrilase. The assay was performed in triplicate at 37 °C in a 96-well microtiter plate. NADH conversion was monitored at 340 nm. Fumaronitrile, succinonitrile, and crotononitrile were degraded after 15 min, thereby defining the threshold (red line) for the required experimental time
To minimize the identification of false positives or enzymes with low activity, two thresholds were defined. During the first 15 min of measurement, total conversion of succinonitrile, fumaronitrile, and crotononitrile by the K22 control nitrilase was recorded and the absorbance decrease at 340 nm was at least 0.75. As K22 nitrilase activity for these substrates was described previously (Kobayashi et al. 1990; Kobayashi et al. 1992b), only reactions active in the first 15 min and with an absorbance decrease of at least 0.75 were considered as positive for substrate conversion and nitrilase activity during subsequent experiments.
Screening of putative metagenome–derived nitrilase-encoding genes and selection of candidates for activity analysis of the corresponding enzymes
Six metagenomes of microbial communities individually obtained from phenylacetonitrile-, succinonitrile-, acetonitrile-, crotononitrile-, 4-hydroxybenzonitrile-, or cyclohexanecarbonitrile-containing enrichment cultures (Egelkamp et al. 2019) served as sources for putative nitrilases. In total, 70 annotated putative nitrilase-encoding genes were identified in these metagenomes (Supplementary Data File S2). In a first step, the sequences of the identified genes and the corresponding deduced proteins were analyzed (Supplementary Data File S2). Initially, to relate the deduced protein sequences with already characterized enzymes, all deduced protein sequences of the metagenome-derived nitrilase candidates were compared with the amino acid sequences of 46 characterized reference nitrilases obtained from SWISS-Prot. This analysis resulted in 13 nitrilase protein clusters (Supplementary Data File S3) and reduced our data set to 60 unique candidates. The first three clusters contained characterized nitrilases obtained from SWISS-Prot as well as our putative metagenome-derived nitrilases with molecular masses ranging from 5.96 to 38.68 kDa (cluster 1), 12.7 to 40.2 kDa (cluster 2), and 10.41 to 38.62 kDa (cluster 3). Clusters 4, 6, 10, 11, 12, and 13 consisted solely of characterized SWISS-Prot-derived nitrilases, whereas the remaining four clusters (5, 7, 8 and 9) included only our putative metagenome-derived enzyme sequences. As the smallest characterized prokaryotic nitrilase originates from Pyrococcus abyssi GE5 and consists of 262 amino acids (Mueller et al. 2006), all putative nitrilases smaller than this were checked for the presence of ribosomal binding sites (RBS) in the corresponding metagenomic DNA region to exclude artificial peptides caused by incomplete metagenomic assembly or misassembly. Finally, 37 candidates were selected for cloning and overexpression in E. coli.
After amplification of the respective genes from metagenomic DNA and sequence verification, frameshift mutations were observed in four cases, leading to their exclusion. In 31 cases, variations resembling point mutations of the metagenome-derived gene sequences were recorded. Probably, the sequencing depth of the metagenomes was not sufficient to distinguish between very similar sequences, leading to the amplification of previously unidentified gene variants. All of these variations were included in subsequent analyses. In total, 26 putative nitrilase-encoding genes were successfully subcloned (Supplementary Data File S2). Analysis of heterologous protein formation revealed production of 13 putative nitrilases by the E. coli host. Although the remaining 13 putative nitrilases could be successfully overexpressed, the corresponding enzymes were not present in the cleared cell-free crude extract. These enzymes were detected in the pellet that remains after cell disruption and clearing the crude extract by centrifugation. These enzymes were not considered for subsequent analysis, as we assumed that these proteins were part of inclusion bodies.
The previously established assay was used to verify nitrilase activity with the 13 putative nitrilase candidates using nine different nitriles as substrate (Table 1). Six of the putative enzymes showed nitrilase activity with at least one nitrile. Most were active with succinonitrile or fumaronitrile. In addition, one of these (Nit09) could act on the aromatic phenylacetonitrile (Fig. 2).
Gene sequences of the six active enzymes and already characterized reference nitrilases were used to calculate a phylogenetic tree (Fig. 3). The phenylacetonitrile-specific nitrilase (Nit09) clustered with arylacetonitrilases, whereas the four nitrilases active with succinonitrile and fumaronitrile (Nit14, Nit28, Nit37, and Nit56) form a branch with an aliphatic nitrilase from Synechocystis sp. PCC6803, and the fumaronitrile-degrading nitrilase (Nit60) most closely resembled with three β-cyano-l-alanine nitrilases.
Characterization of arylacetonitrilase Nit09
Phenylacetonitrile belongs to the class of arylacetonitriles, a subclass of nitriles degraded by the rare group of arylacetonitrilases (EC 3.5.5.5). We chose the phenylacetonitrile-degrading enzyme candidate Nit09 for detailed characterization and verification of enzymatic activity under defined conditions as it represented probably a novel arylacetonitrilase. This assumption was further supported by the clustering of nit09 with other arylacetonitrilases in the previously calculated phylogenetic tree (Fig. 3). The His6-tagged enzyme was produced and purified by immobilized metal ion affinity chromatography. SDS-PAGE analysis of the enzyme preparation confirmed production of a 36 kDa enzyme, which is in accordance with the Nit09 molecular mass deduced from the gene sequence (data not shown). Phenylacetonitrile was used as substrate to further characterize the purified enzyme, since Nit09 showed the ability to degrade phenylacetonitrile in the high-throughput screening.
Enzyme activity was highest at pH 6 with approximately 90% activity being retained between pH 5.5 and 8. At both pH 5 and pH 10.5, the relative activity dropped to 20% (Fig. 4a). Optimum temperature of enzyme activity was at 50 °C, whereas nitrilase activity was reduced to 30% at 60 °C (Fig. 4b). Furthermore, stability of the nitrilase was tested over several weeks under storage conditions at 4 °C. Remaining catalytic activity of > 80% was observed during the first 3 weeks, followed by a slow decline (Fig. 5). After 3 months, 8% of the initial activity remained.
Dependency of Nit09 activity on pH (a) and temperature (b). a Reactions were performed for 2 min at 37 °C in the respective buffers (all 0.1 M final concentration) containing 1.25 μg purified protein and 5 mM phenylacetonitrile. One hundred percent relative activity corresponds to 2.79 U/mg. b Reactions were performed for 2 min at various temperatures using citrate-phosphate buffer (pH 6.0), 1.25 μg purified protein, and 5 mM phenylacetonitrile. One hundred percent relative activity corresponds to 5.68 U/mg
Stability assay with Nit09. Enzyme was stored for 91 days at 4 °C in a buffered system (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 1 mM NaN3, pH 8.0). Reactions were carried out for 2 min at 50 °C using citrate-phosphate buffer (pH 6.0), 1.25 μg purified protein, and 5 mM phenylacetonitrile. One hundred percent relative activity corresponds to 5.68 U/mg
To further examine the substrate spectrum, the Nit09 nitrilase was initially tested with the same substrates used for the high-throughput assay. In this way, the results of the above-described screening assay were verified and phenylacetonitrile was the only one of these substrates converted by Nit09 under the tested conditions (Table 2). To determine activity of the enzyme with structurally related compounds, 13 aromatic nitriles were additionally tested. With four of them (2-phenylbutyronitrile, benzonitrile, 2-naphtylacetonitrile, and cinnamonitrile), nitrilase activity was not detected. With 2-chlorophenylacetonitrile, 3-chlorophenylacetonitrile, and 4-chlorophenylacetonitrile as substrates, only low relative activities (5 to 12%) were recorded compared to phenylacetonitrile. Reduced relative activity was also detected for 3-phenylpropionitrile (16%), 1,4-phenylenediacetonitrile (19%,) mandelonitrile (22%), and 3-indoleacetonitrile (34%). Strikingly, significantly higher relative activity was detected for 3-thiopheneacetonitrile (188%) and 2-thiopheneacetonitril (385%).
To assess the effect of potential inhibitors and solvents on enzyme activity, the activity of the purified enzyme was tested in the presence of different chemicals. For detection of thiol residues essential for enzyme activity, the thiol-binding reagents HgCl2 and AgNO3 were applied to the reaction mixture. The recorded decline of enzyme activity to 1.6 and 2.0%, respectively, indicated that the functionality of Nit09 depends on these residues (Table 3). As the metal-chelating agent EDTA had no significant effect on enzymatic activity, dependence on divalent metal cofactors is not indicated. This was further supported by the lack of any effect when divalent ions such as Mg2+, Fe2+, and Mn2+ were added to the reaction mixtures. Only addition of Zn2+ resulted in a slight relative activity increase to 114%.
Strong inhibitory effects were encountered in the presence of DMSO, acetone, and chloroform even at 5% concentration (Table 4). At this concentration, the nitrilase was less sensitive to methanol, ethanol, isopropanol, and glycerol treatment. At 20% concentration, only glycerol did not abolish enzymatic activity.
Finally, kinetic constants of Nit09 were determined. The reaction rate increased with phenylacetonitrile concentration until a saturation was reached at 6 mM. Between 10 and 50 mM, a rapid decrease of activity was recorded, followed by a slow decline up to 100 mM (Fig. 6a), indicating inhibition by the substrate. Based on these data, kinetic constants were calculated with a non-linear model due to superior precision compared to linear methods like Lineweaver-Burk plots (Cho and Lim 2018). Phenylacetonitrile was converted at 37 °C with a KM of 1.29 mM and Vmax of 13.85 U/mg, reaching the saturation point at 6 mM (Fig. 6b).
Influence of substrate concentrations on the reaction rate of Nit09. Reactions were performed with phenylacetonitrile as substrate under standard conditions at 37 °C. a Steady-state measurement until 100 mM substrate concentration. b Steady-state measurement until saturation point at 6 mM substrate concentration
Discussion
From putative nitrilase sequences to nitrilase function
In this study, a workflow and a fast and simple assay for screening of functional nitrilases was established. In comparison to the previously described method (Vergne-Vaxelaire et al. 2013), it reduces the total screening time from approx. 5 h to approximately 45 min by combining nitrilase reaction and subsequent ammonia conversion in a single reaction. The use of crude extract is less time-consuming compared to methods demanding purified enzymes. Furthermore, the assay system allows continuous measurement of nitrile degradation and provides insights into the efficiency and substrate spectra of the tested enzymes. However, its precision is limited by the GDH as only nitrilase reactions slower than that of GDH can be monitored in detail. In addition, other enzymes in the crude extract might cause a NAD release or regeneration of NAD to NADH, thereby influencing the measurement. To minimize the effect of these factors, additional precautions were taken. Most important is the use of crude extract containing the expression vector without insert as negative control. To further reduce the likelihood of false positives, a time limit for nitrilase reaction was used. Based on the control reactions observed with the well-characterized R. rhodochrous K22 nitrilase (Kobayashi et al. 1990; Kobayashi et al. 1992b), these thresholds were set to 15 min and an absorbance decrease at 340 nm of at least 0.75.
Many of the 70 putative metagenome-derived nitrilase candidate genes were not considered for functional verification. Reason for exclusion was redundancy or problems during expression. In the end, 13 putative nitrilases were screened against 9 different nitriles with 6 of them showing activity for at least one of the nitriles. As nitrilases are known to cluster based on their substrate spectrum (Howden and Preston 2009), the observed substrate specificity was further controlled by calculation of a phylogenetic tree consisting of the 6 active enzymes and characterized nitrilases. The specificity of Nit09 was underlined by its clustering with arylacetonitrilases, as its substrate phenylacetonitrile converted in the initial screening belongs to this class of chemicals. In contrast, the fumaronitrile-specific nitrilase Nit60 contributed to a branch with β-cyano-l-alanine nitrilases. To our knowledge, the three β-cyano-l-alanine nitrilases have not been tested with fumaronitrile or other dinitriles (Howden et al. 2009; Acera et al. 2017), while our test substrates did not contain β-cyano-l-alanine. Thus, we can only assume that Nit60 would show activity with this compound and that the β-cyano-l-alanine nitrilases harbor a so far unknown ability to degrade dinitriles. Furthermore, a cluster consisting of four succinonitrile- and fumaronitrile-degrading nitrilases and a characterized nitrilase from Synechocystis sp. PCC6803 was identified. While the PCC6803-derived nitrilase barely acts on mononitriles, it is highly active for fumaronitrile (Heinemann et al. 2003), further supporting the results of the high-throughput assay. We could show that a combination of targeted sequence data analyses with a high-throughput activity assay assists in the fast identification of functional nitrilase candidates. Another possible application of the high-throughput activity assay is to employ it for the rapid substrate spectrum determination of known or recovered active nitrilases with a variety of different nitriles. The subsequent isolation and verification of the phenylacetonitrile-degrading nitrilase finally proved the reliability of the novel nitrilase screening workflow, which provides a way to successfully narrow down bioinformatically identified enzyme candidates to the interesting functional representatives.
A novel and stable arylacetonitrilase
Arylacetonitrilases (EC 3.5.5.5) are a rare group of enzymes, and to our knowledge, only 12 bacterial nitrilases of this type have been characterized (Table 5). Thus, the phenylacetonitrile-degrading enzyme Nit09 was chosen for further validation of the workflow, as its characterization also contributes to the knowledge on arylacetonitrilases. According to sequence similarity, nit09 is affiliated to Variovorax boronicumulans (Table 1). Nitrile-degrading abilities of this species have been described previously, but phenylacetonitrile degradation has not been mentioned (Nielsen et al. 2007; Zhang et al. 2012; Egelkamp et al. 2017; Sun et al. 2018). The molecular mass of Nit09 (36 kDa) is close to molecular masses of other known arylacetonitrilases such as the enzymes from Pseudomonas sp. UW4 (33 kDa; Duca et al. 2014), Bradyrhizobium japonicum USDA110 (37 kDa; Zhu et al. 2007a), or P. fluorescens EBC191 (38 kDa; Kiziak et al. 2005). In contrast, the broad pH range of Nit09 activity is not that common for nitrilases of this type, including the enzymes from Alcaligenes faecalis JM3 (Nagasawa et al. 1990) and A. faecalis ATCC 8750 (Yamamoto et al. 1992). In addition, Nit09 exhibits an unusual long-term stability with 80% remaining activity after 3 weeks of incubation at 4 °C; in contrast, the arylacetonitrilases from A. faecalis JM3 and P. fluorescens EBC191 lose up to 80% activity in 10 days.
During initial screening, minor degradation of other nitriles was observed in the crude extract containing Nit09. None of these substrates was degraded by the purified enzyme, underlining the importance of the 15-min threshold to avoid identification of false positive candidates.
Additional tests with substrates structurally related to phenylacetonitrile provided a deeper insight into specificity of the nitrilase. Weak activity on non-arylacetonitriles was only recorded for 3-phenypropionitrile, demonstrating the importance of an acetonitrile-like residue attached to the aryl structure for catalytic function. Halogenic ortho, meta, and para substitutions at the benzyl residue led compared with phenylacetonitrile as substrate to a strong reduction of enzymatic activity. This behavior is not common for other arylacetonitrilases such as the enzymes from A. faecalis JM3 (Nagasawa et al. 1990), A. faecalis ATCC 8750 (Yamamoto et al. 1992), and P. putida MTCC 5110 (Banerjee et al. 2006), as in all tested cases at least one type of substitution led to increased activity. Interestingly, 1,4-phenylenediacetonitrile was converted with 19% relative activity, indicating an interference of halogens with the active center. Reduced activity was also observed for the plant hormone predecessor 3-indoleacetonitrile. The larger aromatic structure seems to cause general steric problems, as this is also the case for other arylacetonitrilases such as the enzymes from A. faecalis JM3 (Nagasawa et al. 1990), Burkholderia cenocepacia J2315 (Wang et al. 2013), P. fluorescens EBC191 (Kiziak et al. 2005), and P. putida MTCC 5110 (Banerjee et al. 2006) exhibited reduced activity with this compound. In contrast, the heteroaromatic structure found in thiophene results in increased enzymatic activity compared to phenylacetonitrile, probably due to the assistance of the sulfur in the nucleophilic attack performed by nitrilases (Ramteke et al. 2013). Furthermore, an effect of the position of the heteroatom was recorded. 2-Thiopheneacetonitrile was converted more than twice as efficient as 3-thiopheneacetonitrile by Nit09. This strong influence of the heteroatom is typical for arylacetonitrilases and can lead to more than a tenfold difference in enzymatic activity (Nagasawa et al. 1990; Yamamoto et al. 1992; Kiziak et al. 2005).
Nit09 was strongly inhibited by the thiol-specific reagents AgNO3 and HgCl2. Most likely, thiol groups build an important part of the active site, which is in line with the proposed Cys-Glu-Lys catalytic triad of nitrilases (Fernandes et al. 2006). Most arylacetonitrilases such as the enzymes from A. faecalis JM3 (Nagasawa et al. 1990), A. faecalis ATCC 8750 (Yamamoto et al. 1992), and A. faecalis ZJUTB10 (Liu et al. 2011) are also inhibited by other heavy metal ions like Cu2+ or Co2+, whereas the activity of Nit09 was almost unaffected by the presence of these ions. Interestingly, an increased activity was detected in the presence of ZnCl2. In general, nitrilase activities are independent of metal cofactors (Gong et al. 2012). This was supported by the lack of Nit09 activity inhibition by the presence of the chelating agents EDTA and sodium azide. Thus, a different mechanism like enhanced protein stability in the presence of Zn2+ might explain the detected activity increase of Nit09.
The characterization of arylacetonitrilase Nit09 provided additional knowledge on this rare group of nitrilases. The substrate range is common for this enzyme type, but the combination of a low pH optimum, a high temperature optimum, and an unusual long-term stability with a pronounced resistance against most metal ions is exceptional. Arylacetonitrilases are industrially interesting biocatalysts as they are currently used for the large-scale conversion of mandelonitrile to mandelic acid (Gong et al. 2012). Thus, identification of new enzymes of this type might open new production routes. Interesting in this respect is the conversion of phenylacetonitrile to phenylacetic acid, which is used among others for the production of penicillin G (Ziemons et al. 2017). The new insights obtained by identification and analysis of Nit09 contribute to the understanding of the sequence function relationship of arylacetonitrilases and thus to the development of better or novel nitrilase biocatalysts.
References
Acera F, Carmona MI, Castillo F, Quesada A, Blasco R (2017) A cyanide-induced 3-cyanoalanine nitrilase in the cyanide-assimilating bacterium Pseudomonas pseudoalcaligenes strain CECT 5344. Appl Environ Microbiol 83:e00089–e00017. https://doi.org/10.1128/AEM.00089-17
Almatawah QA, Cowan DA (1999) Thermostable nitrilase catalysed production of nicotinic acid from 3-cyanopyridine. Enzym Microb Technol 25:718–724. https://doi.org/10.1016/S0141-0229(99)00104-0
Ambler RP, Auffret AD, Clarke PH (1987) The amino acid sequence of the aliphatic amidase from Pseudomonas aeruginosa. FEBS Lett 215:285–290. https://doi.org/10.1016/0014-5793(87)80163-1
Bairoch A, Apweiler R (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res 28:45–48. https://doi.org/10.1093/nar/28.1.45
Banerjee A, Sharma R, Banerjee UC (2003) A rapid and sensitive fluorometric assay method for the determination of nitrilase activity. Biotechnol Appl Biochem 37:289–293. https://doi.org/10.1042/BA20020106
Banerjee A, Kaul P, Banerjee UC (2006) Purification and characterization of an enantioselective arylacetonitrilase from Pseudomonas putida. Arch Microbiol 184:407–418. https://doi.org/10.1007/s00203-005-0061-9
Banerjee A, Dubey S, Kaul P, Barse B, Piotrowski M, Banerjee UC (2009) Enantioselective nitrilase from Pseudomonas putida: cloning, heterologous expression, and bioreactor studies. Mol Biotechnol 41:35–41. https://doi.org/10.1007/s12033-008-9094-z
Bayer S, Birkemeyer C, Ballschmiter M (2011) A nitrilase from a metagenomic library acts regioselectively on aliphatic dinitriles. Appl Microbiol Biotechnol 89:91–98. https://doi.org/10.1007/s00253-010-2831-9
Bhatia SK, Mehta PK, Bhatia RK, Bhalla TC (2014) Purification and characterization of arylacetonitrile-specific nitrilase of Alcaligenes sp. MTCC 10675. Biotechnol Appl Biochem 61:459–465. https://doi.org/10.1002/bab.1192
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Bui K, Maestracci M, Thiery A, Arnaud A, Galzy P (1984) A note on the enzymic action and biosynthesis of a nitrile-hydratase from a Brevibacterium sp. J Appl Bacteriol 57:183–190. https://doi.org/10.1111/j.1365-2672.1984.tb02373.x
Cho Y-S, Lim H-S (2018) Comparison of various estimation methods for the parameters of Michaelis-Menten equation based on in vitro elimination kinetic simulation data. Transl Clin Pharmacol 26:39-47. https://doi.org/10.12793/tcp.2018.26.1.39
Conn EE (1979) Biosynthesis of cyanogenic glycosides. Naturwissenschaften 66:28–34. https://doi.org/10.1007/BF00369352
Crum MA-N, Park JM, Mulelu AE, Sewell BT, Benedik MJ (2015) Probing C-terminal interactions of the Pseudomonas stutzeri cyanide-degrading CynD protein. Appl Microbiol Biotechnol 99:3093–3102. https://doi.org/10.1007/s00253-014-6335-x
Duca D, Rose DR, Glick BR (2014) Characterization of a nitrilase and a nitrile hydratase from Pseudomonas sp. strain UW4 that converts indole-3-acetonitrile to indole-3-acetic acid. Appl Environ Microbiol 80:4640–4649. https://doi.org/10.1128/AEM.00649-14
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Egelkamp R, Schneider D, Hertel R, Daniel R (2017) Nitrile-degrading bacteria isolated from compost. Front Environ Sci 5:56. https://doi.org/10.3389/fenvs.2017.00056
Egelkamp R, Zimmermann T, Schneider D, Hertel R, Daniel R (2019) Impact of nitriles on bacterial communities. Front Environ Sci 7:103. https://doi.org/10.3389/fenvs.2019.00103
Fawcett JK, Scott JE (1960) A rapid and precise method for the determination of urea. J Clin Pathol 13:156–159. https://doi.org/10.1136/jcp.13.2.156
Fernandes BCM, Mateo C, Kiziak C, Chmura A, Wacker J, van Rantwijk F, Stolz A, Sheldon RA (2006) Nitrile hydratase activity of a recombinant nitrilase. Adv Synth Catal 348:2597–2603. https://doi.org/10.1002/adsc.200600269
Gong J-S, Lu Z-M, Li H, Shi J-S, Zhou Z-M, Xu Z-H (2012) Nitrilases in nitrile biocatalysis: recent progress and forthcoming research. Microb Cell Factories 11:142. https://doi.org/10.1186/1475-2859-11-142
Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, Jahn D (2005) JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 33:W526–W531. https://doi.org/10.1093/nar/gki376
Guzman L-M, Belin D, Carson MJ, Beckwith J (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. https://doi.org/10.1128/jb.177.14.4121-4130.1995
Heinemann U, Engels D, Bürger S, Kiziak C, Mattes R, Stolz A (2003) Cloning of a nitrilase gene from the cyanobacterium Synechocystis sp. strain PCC6803 and heterologous expression and characterization of the encoded protein. Appl Environ Microbiol 69:4359–4366. https://doi.org/10.1128/AEM.69.8.4359-4366.2003
Howden AJM, Preston GM (2009) Nitrilase enzymes and their role in plant-microbe interactions. Microb Biotechnol 2:441–451. https://doi.org/10.1111/j.1751-7915.2009.00111.x
Howden AJM, Harrison CJ, Preston GM (2009) A conserved mechanism for nitrile metabolism in bacteria and plants. Plant J 57:243–253. https://doi.org/10.1111/j.1365-313X.2008.03682.x
Huang Y, Niu B, Gao Y, Fu L, Li W (2010) CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682. https://doi.org/10.1093/bioinformatics/btq003
Jallageas J-C, Arnaud A, Galzy P (1980) Bioconversion of nitriles and their applications. In: Fiechter A (ed) Advances in biochemical engineering, 1st edn. Springer Berlin Heidelberg, Berlin, pp 1–32
Jandhyala D, Berman M, Meyers PR, Sewell BT, Willson RC, Benedik MJ (2003) CynD, the cyanide dihydratase from Bacillus pumilus: gene cloning and structural studies. Appl Environ Microbiol 69:4794–4805. https://doi.org/10.1128/AEM.69.8.4794-4805.2003
Kaplan A, Chaney AL, Lynch RL, Meites S (1965) Urea nitrogen and urinary ammonia. In: Meites S (ed) Standard methods of clinical chemistry, 1st edn. Academic Press, New York, pp 245–256
Kiziak C, Conradt D, Stolz A, Mattes R, Klein J (2005) Nitrilase from Pseudomonas fluorescens EBC191: cloning and heterologous expression of the gene and biochemical characterization of the recombinant enzyme. Microbiology 151:3639–3648. https://doi.org/10.1099/mic.0.28246-0
Kobayashi M, Yanaka N, Nagasawa T, Yamada H (1990) Purification and characterization of a novel nitrilase of Rhodococcus rhodochrous K22 that acts on aliphatic nitriles. J Bacteriol 172:4807–4815. https://doi.org/10.1128/jb.172.9.4807-4815.1990
Kobayashi M, Komeda H, Yanaka N, Nagasawa T, Yamada H (1992a) Nitrilase from Rhodococcus rhodochrous J1: sequencing and overexpression of the gene and identification of an essential cysteine residue. J Biol Chem 267:20746–20751
Kobayashi M, Yanaka N, Nagasawa T, Yamada H (1992b) Primary structure of an aliphatic nitrile-degrading enzyme, aliphatic nitrilase, from Rhodococcus rhodochrous K22 and expression of its gene and identification of its active site residue. Biochemistry 31:9000–9007. https://doi.org/10.1021/bi00152a042
Kobayashi M, Izui H, Nagasawa T, Yamada H (1993) Nitrilase in biosynthesis of the plant hormone indole-3-acetic acid from indole-3-acetonitrile: cloning of the Alcaligenes gene and site-directed mutagenesis of cysteine residues. Proc Natl Acad Sci U S A 90:247–251. https://doi.org/10.1073/pnas.90.1.247
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Lévy-Schil S, Soubrier F, Crutz-Le Coq AM, Faucher D, Crouzet J, Pétré D (1995) Aliphatic nitrilase from a soil-isolated Comamonas testosteroni sp.: gene cloning and overexpression, purification and primary structure. Gene 161:15–20. https://doi.org/10.1016/0378-1119(95)00242-X
Liu ZQ, Dong LZ, Cheng F, Xue YP, Wang YS, Ding JN, Zheng YG, Shen YC (2011) Gene cloning, expression, and characterization of a nitrilase from Alcaligenes faecalis ZJUTB10. J Agric Food Chem 59:11560–11570. https://doi.org/10.1021/jf202746a
Martínková L, Uhnáková B, Pátek M, Nešvera J, Křen V (2009) Biodegradation potential of the genus Rhodococcus. Environ Int 35:162–177. https://doi.org/10.1016/j.envint.2008.07.018
Mueller P, Egorova K, Vorgias CE, Boutou E, Trauthwein H, Verseck S, Antranikian G (2006) Cloning, overexpression, and characterization of a thermoactive nitrilase from the hyperthermophilic archaeon Pyrococcus abyssi. Protein Expr Purif 47:672–681. https://doi.org/10.1016/j.pep.2006.01.006
Nagasawa T, Mauger J, Yamada H (1990) A novel nitrilase, arylacetonitrilase, of Alcaligenes faecalis JM3. Purification and characterization. Eur J Biochem 194:765–772. https://doi.org/10.1111/j.1432-1033.1990.tb19467.x
Nei M, Kumar S (2000) Molecular evolution and phylogenetics, 1st edn. Oxford University Press, New York
Nielsen MKK, Holtze MS, Svensmark B, Juhler RK (2007) Demonstrating formation of potentially persistent transformation products from the herbicides bromoxynil and ioxynil using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Pest Manag Sci 63:141–149. https://doi.org/10.1002/ps.1311
Panova A, Mersinger LJ, Liu Q, Foo T, Roe DC, Spillan WL, Sigmund AE, Ben-Bassat A, Wagner LW, O’Keefe DP, Wu S, Petrillo KL, Payne MS, Breske ST, Gallagher FG, DiCosimo R (2007) Chemoenzymatic synthesis of glycolic acid. Adv Synth Catal 349:1462–1474. https://doi.org/10.1002/adsc.200700061
Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GSA, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R, Dodson RJ, Durkin AS, Brinkac LM, Daugherty SC, Sullivan SA, Rosovitz MJ, Gwinn ML, Zhou L, Schneider DJ, Cartinhour SW, Nelson WC, Weidman J, Watkins K, Tran K, Khouri H, Pierson EA, Pierson LS, Thomashow LS, Loper JE (2005) Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol 23:873–878. https://doi.org/10.1038/nbt1110
Piotrowski M (2008) Primary or secondary? Versatile nitrilases in plant metabolism. Phytochemistry 69:2655–2667. https://doi.org/10.1016/j.phytochem.2008.08.020
Ramteke PW, Maurice NG, Joseph B, Wadher BJ (2013) Nitrile-converting enzymes: an eco-friendly tool for industrial biocatalysis. Biotechnol Appl Biochem 60:459–481. https://doi.org/10.1002/bab.1139
Reisinger C, van Assema F, Schürmann M, Hussain Z, Remler P, Schwab H (2006) A versatile colony assay based on NADH fluorescence. J Mol Catal B Enzym 39:149–155. https://doi.org/10.1016/j.molcatb.2006.01.014
Ress-Löschke M, Hauer B, Mattes R, Engels D (2001) Nitrilase from Rhodococcus rhodochrous NCIMB 11216. BASF AG, German patent number DE10010149
Riabchenko L, Podcherniaev D, Kotlova E, Yanenko A (2006) Cloning the amidase gene from Rhodococcus rhodochrous M8 and its expression in Escherichia coli. Genetika 42:1075–1082. https://doi.org/10.1134/S1022795406080060
Ritz C, Baty F, Streibig JC, Gerhard D (2015) Dose-response analysis using R. PLoS One 10:e0146021. https://doi.org/10.1371/journal.pone.0146021
Robertson DE, Chaplin J, DeSantis G, Podar M, Madden M, Chi E, Richardson T, Milan A, Miller M, Weiner DP, Wong K, McQuaid J, Farwell B, Preston L, Tan X, Snead M, Keller M, Mathur E, Kretz PL, Burk MJ, Short JM (2004) Exploring nitrilase sequence space for enantioselective catalysis. Appl Environ Microbiol 70:2429–2436. https://doi.org/10.1128/AEM.70.4.2429-2436.2004
Rustler S, Stolz A (2007) Isolation and characterization of a nitrile hydrolysing acidotolerant black yeast - Exophiala oligosperma R1. Appl Microbiol Biotechnol 75:899–908. https://doi.org/10.1007/s00253-007-0890-3
Seffernick JL, Samanta SK, Louie TM, Wackett LP, Subramanian M (2009) Investigative mining of sequence data for novel enzymes: a case study with nitrilases. J Biotechnol 143:17–26. https://doi.org/10.1016/j.jbiotec.2009.06.004
Shaw NM, Robins KT, Kiener A (2003) Lonza: 20 years of biotransformations. Adv Synth Catal 345:425–435. https://doi.org/10.1002/adsc.200390049
Silby MW, Cerdeño-Tárraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, Zhang X-X, Moon CD, Gehrig SM, Godfrey SAC, Knight CG, Malone JG, Robinson Z, Spiers AJ, Harris S, Challis GL, Yaxley AM, Harris D, Seeger K, Murphy L, Rutter S, Squares R, Quail MA, Saunders E, Mavromatis K, Brettin TS, Bentley SD, Hothersall J, Stephens E, Thomas CM, Parkhill J, Levy SB, Rainey PB, Thomson NR (2009) Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol 10:R51. https://doi.org/10.1186/gb-2009-10-5-r51
Soares Bragança CR, Dooley-Cullinane T-M, O’Reilly C, Coffey L (2017) Applying functional metagenomics to search for novel nitrile-hydrolyzing enzymes using environmental samples. Biomater Tissue Technol 1:1000108. https://doi.org/10.15761/BTT.1000108
Sun H, Gao W, Fan H, Wang H, Wei D (2015) Cloning, purification and evaluation of the enzymatic properties of a novel arylacetonitrilase from Luminiphilus syltensis NOR5-1B: a potential biocatalyst for the synthesis of mandelic acid and its derivatives. Biotechnol Lett 37:1655–1661. https://doi.org/10.1007/s10529-015-1830-4
Sun S-L, Yang W-L, Fang W-W, Zhao Y-X, Guo L, Dai Y-J (2018) The plant growth-promoting rhizobacterium Variovorax boronicumulans CGMCC 4969 regulates the level of indole-3-acetic acid synthesized from indole-3-acetonitrile. Appl Environ Microbiol 84:e00298–e00218. https://doi.org/10.1128/AEM.00298-18
Vaughan PA, Knowles CJ, Cheetham PSJ (1989) Conversion of 3-cyanopyridine to nicotinic acid by Nocardia rhodochrous LL100-21. Enzym Microb Technol 11:815–823. https://doi.org/10.1016/0141-0229(89)90055-0
Vergne-Vaxelaire C, Bordier F, Fossey A, Besnard-Gonnet M, Debard A, Mariage A, Pellouin V, Perret A, Petit J-L, Stam M, Salanoubat M, Weissenbach J, De Berardinis V, Zaparucha A (2013) Nitrilase activity screening on structurally diverse substrates: providing biocatalytic tools for organic synthesis. Adv Synth Catal 355:1763–1779. https://doi.org/10.1002/adsc.201201098
Wang H, Sun H, Wei D (2013) Discovery and characterization of a highly efficient enantioselective mandelonitrile hydrolase from Burkholderia cenocepacia J2315 by phylogeny-based enzymatic substrate specificity prediction. BMC Biotechnol 13:14. https://doi.org/10.1186/1472-6750-13-14
Wibberg D, Luque-Almagro VM, Igeño MI, Bremges A, Roldán MD, Merchán F, Sáez LP, Guijo MI, Manso MI, Macías D, Cabello P, Becerra G, Ibáñez MI, Carmona MI, Escribano MMP, Castillo F, Sczyrba A, Moreno-Vivián C, Blasco R, Pühler A, Schlüter A (2014) Complete genome sequence of the cyanide-degrading bacterium Pseudomonas pseudoalcaligenes CECT5344. J Biotechnol 175:67–68. https://doi.org/10.1016/j.jbiotec.2014.02.004
Williamson DS, Dent KC, Weber BW, Varsani A, Frederick J, Thuku RN, Cameron RA, van Heerden JH, Cowan DA, Sewell BT (2010) Structural and biochemical characterization of a nitrilase from the thermophilic bacterium, Geobacillus pallidus RAPc8. Appl Microbiol Biotechnol 88:143–153. https://doi.org/10.1007/s00253-010-2734-9
Yamamoto K, Fujimatsu I, Komatsu KI (1992) Purification and characterization of the nitrilase from Alcaligenes faecalis ATCC 8750 responsible for enantioselective hydrolysis of mandelonitrile. J Ferment Bioeng 73:425–430. https://doi.org/10.1016/0922-338X(92)90131-D
Yazbeck DR, Durao PJ, Xie Z, Tao J (2006) A metal ion-based method for the screening of nitrilases. J Mol Catal B Enzym 39:156–159. https://doi.org/10.1016/j.molcatb.2006.01.036
Zhang Z-J, Pan J, Liu J-F, Xu J-H, He Y-C, Liu Y-Y (2011a) Significant enhancement of (R)-mandelic acid production by relieving substrate inhibition of recombinant nitrilase in toluene-water biphasic system. J Biotechnol 152:24–29. https://doi.org/10.1016/j.jbiotec.2011.01.013
Zhang Z-J, Xu J-H, He Y-C, Ouyang L-M, Liu Y-Y (2011b) Cloning and biochemical properties of a highly thermostable and enantioselective nitrilase from Alcaligenes sp. ECU0401 and its potential for (R)-(-)-mandelic acid production. Bioprocess Biosyst Eng 34:315–222. https://doi.org/10.1007/s00449-010-0473-z
Zhang H-J, Zhou Q-W, Zhou G-C, Cao Y-M, Dai Y-J, Ji W-W, Shang G-D, Yuan S (2012) Biotransformation of the neonicotinoid insecticide thiacloprid by the bacterium Variovorax boronicumulans strain J1 and mediation of the major metabolic pathway by nitrile hydratase. J Agric Food Chem 60:153–159. https://doi.org/10.1021/jf203232u
Zhu D, Mukherjee C, Biehl ER, Hua L (2007a) Discovery of a mandelonitrile hydrolase from Bradyrhizobium japonicum USDA110 by rational genome mining. J Biotechnol 129:645–650. https://doi.org/10.1016/j.jbiotec.2007.02.001
Zhu Q, Fan A, Wang Y, Zhu X, Wang Z, Wu M, Zheng Y (2007b) Novel sensitive high-throughput screening strategy for nitrilase-producing strains. Appl Environ Microbiol 73:6053–6057. https://doi.org/10.1128/AEM.01089-07
Ziemons S, Koutsantas K, Becker K, Dahlmann T, Kück U (2017) Penicillin production in industrial strain Penicillium chrysogenum P2niaD18 is not dependent on the copy number of biosynthesis genes. BMC Biotechnol 17:16. https://doi.org/10.1186/s12896-017-0335-8
Acknowledgments
We thank Patrick Kloskowski and Mechthild Bömeke for providing technical assistance.
Funding
Open Access funding provided by Projekt DEAL. The work of Richard Egelkamp was supported by the “Fonds der Chemischen Industrie im Verband der Chemischen Industrie e.V.”.
Author information
Authors and Affiliations
Contributions
RE, RH, and RD conceived the study. IF performed the experiments. RE and IF analyzed the data. All authors interpreted the results, wrote the manuscript, and reviewed the final version of the manuscript.
Corresponding author
Ethics declarations
This article does not contain any studies with human participants or animals performed by any of the authors.
Disclaimer
The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
This article contains supplementary material. The supplementary data files (Data File S1, Data File S2 and Data File S3), Table S1 and Figure S1 are given in the following files: Supplementary Data_File_S1.fasta, Supplementary Data_File_S2.xlsx, Supplementary Data_File_S3.txt and Supplementary_Figure_S1_and_Table_S1.pdf.
ESM 1
(FASTA 39 kb)
ESM 2
(XLSX 56 kb)
ESM 3
(TXT 5 kb)
ESM 4
(PDF 220 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Egelkamp, R., Friedrich, I., Hertel, R. et al. From sequence to function: a new workflow for nitrilase identification. Appl Microbiol Biotechnol 104, 4957–4970 (2020). https://doi.org/10.1007/s00253-020-10544-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10544-9