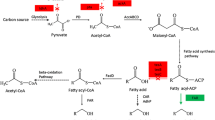
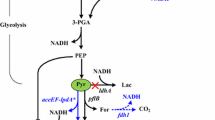
Abstract
Microbial-produced branched-chain higher alcohols (BCHAs), such as isopropanol, isobutanol, and isopentanol in Escherichia coli, have emerged as promising alternative biofuels under development. Elucidating and improving the tolerance of E. coli to BCHAs are important issues for microbial production of BCHAs due to their physiological inhibitory effect. Previous works aimed at understanding the genetic basis of E. coli tolerance to BCHAs with a comparative genome, reverse engineering, or transcriptome approach have gained some important insights into the mechanism of tolerance. However, investigation on BCHA tolerance from the whole-genomic, transcriptomic, and metabolic levels via a systematic approach has not yet been completely elucidated. Here, in this study, genomic, transcriptomic, and 13C-metabolic flux analyses (13C-MFA) of an evolved E. coli strain adapted to BCHA tolerance were conducted. Genome mutation of negative regulation factor (rssB, acrB, and clpX) of RpoS level suggested upregulation of RpoS activity in BCHA tolerance of E. coli. From a more detailed perspective, enhanced energy metabolism was observed to be the main characteristic of E. coli strain tolerant to BCHAs. Enhanced energy metabolism has been achieved through several routes, which included redistribution of the central carbon metabolism, upregulation of the energy generation machinery, and facilitating the operation of electron transferring chain. Evidence of multiple solutions of genotype modification toward BCHA tolerance was also revealed through comparative analysis of previous works from different groups.







Similar content being viewed by others
References
Anto nino Baez K-MC, Liao JC (2011) High-flux isobutanol production using engineered Escherichia coli : a bioreactor study with in situ product removal. Appl Microbiol Biotechnol 90:1681–1690. https://doi.org/10.1007/s00253-011-3173-y
Antoniewicz MR (2018) A guide to (13)C metabolic flux analysis for the cancer biologist. Exp Mol Med 50(4):19. https://doi.org/10.1038/s12276-018-0060-y
Atsumi S, Hanai T, Liao JC (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451(7174):86–9. https://doi.org/10.1038/nature06450
Atsumi S, Wu TY, Machado IM, Huang WC, Chen PY, Pellegrini M, Liao JC (2010) Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol 6:449. https://doi.org/10.1038/msb.2010.98
Barrick JE, Colburn G, Deatherage DE, Traverse CC, Strand MD, Borges JJ, Knoester DB, Reba A, Meyer AG (2014) Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics 15:1039. https://doi.org/10.1186/1471-2164-15-1039
Battesti A, Majdalani N, Gottesman S (2011) The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. https://doi.org/10.1146/annurev-micro-090110-102946
Brynildsen MP, Liao JC (2009) An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol Syst Biol 5:277. https://doi.org/10.1038/msb.2009.34
Ceccato-Antonini SR, da Silva PC (2002) Hyphal-like extension and pseudohyphal formation in industrial strains of yeasts induced by isoamyl alcohol. Braz J of Microbioly 33(3):209–212
Chen Z, Liu D (2016) Toward glycerol biorefinery: metabolic engineering for the production of biofuels and chemicals from glycerol. Biotechnol biofuels 9:205. https://doi.org/10.1186/s13068-016-0625-8
Crook N, Sun J, Morse N, Schmitz A, Alper HS (2016) Identification of gene knockdown targets conferring enhanced isobutanol and 1-butanol tolerance to Saccharomyces cerevisiae using a tunable RNAi screening approach. Appl Microbiol Biotechnol 100(23):10005–10018. https://doi.org/10.1007/s00253-016-7791-2
Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, Keasling JD (2013) Engineering dynamic pathway regulation using stress-response promoters. Nat Biotechnol 31(11):1039–1046. https://doi.org/10.1038/nbt.2689
Deatherage DE, Barrick JE (2014) Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. https://doi.org/10.1007/978-1-4939-0554-6_12
Dunlop M (2011) Engineering microbes for tolerance to next-generation biofuels. Biotechnol biofuels 4(1):32
Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A (2011) Engineering microbial biofuel tolerance and export using efflux pumps. Mol Syst Biol 7:487. https://doi.org/10.1038/msb.2011.21
Eleutherio E, Panek A, De Mesquita JF, Trevisol E, Magalhaes R (2015) Revisiting yeast trehalose metabolism. Curr Genet 61(3):263–274. https://doi.org/10.1007/s00294-014-0450-1
Feng X, Tang KH, Blankenship RE, Tang YJ (2010) Metabolic flux analysis of the mixotrophic metabolisms in the green sulfur bacterium chlorobaculum tepidum. J Biol Chem 285(50):39544–39550. https://doi.org/10.1074/jbc.M110.162958
Hauser M, Horn P, Tournu H, Hauser NC, Hoheisel JD, Brown AJ, Dickinson JR (2007) A transcriptome analysis of isoamyl alcohol-induced filamentation in yeast reveals a novel role for Gre2p as isovaleraldehyde reductase. FEMS Yeast Res 7(1):84–92. https://doi.org/10.1111/j.1567-1364.2006.00151.x
Hayakawa K, Matsuda F, Shimizu H (2018) (13)C-metabolic flux analysis of ethanol-assimilating Saccharomyces cerevisiae for S-adenosyl-L-methionine production. Microb Cell Fact 17(1):82. https://doi.org/10.1186/s12934-018-0935-6
He L, Wu SG, Zhang M, Chen Y, Tang YJ (2016) WUFlux: an open-source platform for 13C metabolic flux analysis of bacterial metabolism. BMC Bioinformatics 17(1):444–447. https://doi.org/10.1186/s12859-016-1314-0
He L, Xiao Y, Gebreselassie N, Zhang F, Antoniewiez MR, Tang YJ, Peng L (2014) Central metabolic responses to the overproduction of fatty acids in Escherichia coli based on 13C-metabolic flux analysis. Biotechnol Bioeng 111(3):575–585. https://doi.org/10.1002/bit.25124
Horinouchi T, Sakai A, Kotani H, Tanabe K, Furusawa C (2017) Improvement of isopropanol tolerance of Escherichia coli using adaptive laboratory evolution and omics technologies. J Biotechnol 255:47–56. https://doi.org/10.1016/j.jbiotec.2017.06.408
Inokuma K, Liao JC, Okamoto M, Hanai T (2010) Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping. J Biosci Bioeng 110(6):696–701. https://doi.org/10.1016/j.jbiosc.2010.07.010
Iordachescu M, Imai R (2008) Trehalose biosynthesis in response to abiotic stresses. J Integr Plant Biol 50(10):1223–1229. https://doi.org/10.1111/j.1744-7909.2008.00736.x
Jin H, Chen L, Wang J, Zhang W (2014) Engineering biofuel tolerance in non-native producing microorganisms. Biotechnol Adv 32(2):541–548. https://doi.org/10.1016/j.biotechadv.2014.02.001
Jozefczuk S, Klie S, Catchpole G, Szymanski J, Cuadros-Inostroza A, Steinhauser D, Selbig J, Willmitzer L (2010) Metabolomic and transcriptomic stress response of Escherichia coli. Mol Syst Biol 6:364. https://doi.org/10.1038/msb.2010.18
Kern K, Nunn CD, Pichova A, Dickinson JR (2004) Isoamyl alcohol-induced morphological change in Saccharomyces cerevisiae involves increases in mitochondria and cell wall chitin content. FEMS Yeast Res 5(1):43–49. https://doi.org/10.1016/j.femsyr.2004.06.011
Lee SK, Chou H, Ham TS, Lee TS, Keasling JD (2008) Metabolic engineering of microorganisms for biofuels production: from bugs to synthetic biology to fuels. Curr Opin Biotechnol 19(6):556–563. https://doi.org/10.1016/j.copbio.2008.10.014
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25(15):1966–1967. https://doi.org/10.1093/bioinformatics/btp336
Luan G, Cai Z, Li Y, Ma Y (2013) Genome replication engineering assisted continuous evolution (GREACE) to improve microbial tolerance for biofuels production. Biotechnol Biofuels 6(1):137. https://doi.org/10.1186/1754-6834-6-137
Minty JJ, Lesnefsky AA, Lin F, Chen Y, Zaroff TA, Veloso AB, Xie B, McConnell CA, Ward RJ, Schwartz DR, Rouillard JM, Gao Y, Gulari E, Lin XN (2011) Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb Cell Factories 10:18. https://doi.org/10.1186/1475-2859-10-18
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628. https://doi.org/10.1038/nmeth.1226
Mukhopadhyay A (2015) Tolerance engineering in bacteria for the production of advanced biofuels and chemicals. Trends Microbiol 23(8):498–508. https://doi.org/10.1016/j.tim.2015.04.008
Niu FX, He X, Wu YQ, Liu JZ (2018) Enhancing production of pinene in Escherichia coli by using a combination of tolerance, evolution, and modular co-culture engineering. Front Microbiol 9:1623. https://doi.org/10.3389/fmicb.2018.01623
Otoupal PB, Chatterjee A (2018) CRISPR gene perturbations provide insights for improving bacterial biofuel tolerance. Front Bioeng Biotechnol 6:122. https://doi.org/10.3389/fbioe.2018.00122
Peng Z, Cheng Y, Tan BC-M, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, Guo J, Dong Z, Liang Y, Bao L, Wang J (2012) Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotech 30(3):253–260. http://www.nature.com/nbt/journal/v30/n3/abs/nbt.2122.html#supplementary-information
Rau MH, Calero P, Lennen RM, Long KS, Nielsen AT (2016) Genome-wide Escherichia coli stress response and improved tolerance towards industrially relevant chemicals. Microb Cell Factories 15(1):176. https://doi.org/10.1186/s12934-016-0577-5
Ren S, Peng Z, Mao J-H, Yu Y, Yin C, Gao X, Cui Z, Zhang J, Yi K, Xu W, Chen C, Wang F, Guo X, Lu J, Yang J, Wei M, Tian Z, Guan Y, Tang L, Xu C, Wang L, Gao X, Tian W, Wang J, Yang H, Wang J, Sun Y (2012) RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res 22(5):806–821. http://www.nature.com/cr/journal/v22/n5/suppinfo/cr201230s1.html
Roell GW, Zha J, Carr RR, Koffas MA, Fong SS, Tang YJ (2019) Engineering microbial consortia by division of labor. Microb Cell Factories 18(1):35. https://doi.org/10.1186/s12934-019-1083-3
Rutherford BJ, Dahl RH, Price RE, Szmidt HL, Benke PI, Mukhopadhyay A, Keasling JD (2010) Functional genomic study of exogenous n-butanol stress in Escherichia coli. Appl Environ Microbiol 76(6):1935–1945. https://doi.org/10.1128/AEM.02323-09
Sauer U, Hatzimanikatis V, Bailey JE, Hochuli M, Szyperski T, Wuthrich K (1997) Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat Biotechnol 15(5):448–452. https://doi.org/10.1038/nbt0597-448
Schwechheimer SK, Becker J, Wittmann C (2018) Towards better understanding of industrial cell factories: novel approaches for (13)C metabolic flux analysis in complex nutrient environments. Curr Opin Biotechnol 54:128–137. https://doi.org/10.1016/j.copbio.2018.07.001
Siu Y, Fenno J, Lindle JM, Dunlop MJ (2018) Design and selection of a synthetic feedback loop for optimizing biofuel tolerance. ACS Synth Biol 7(1):16–23. https://doi.org/10.1021/acssynbio.7b00260
Stephanopoulos G (1999) Metabolic fluxes and metabolic engineering. Metab Eng 1(1):1–11. https://doi.org/10.1006/mben.1998.0101
Szyperski T (1995) Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids. An efficient analytical tool to investigate intermediary metabolism. Eur J Biochem / FEBS 232(2):433–448
Turner TL, Kim H, Kong II, Liu JJ, Zhang GC, Jin YS (2016) Engineering and evolution of Saccharomyces cerevisiae to produce biofuels and chemicals. Adv Biochem Eng Biotechnol. https://doi.org/10.1007/10_2016_22
Turner WJ, Dunlop MJ (2015) Trade-offs in improving biofuel tolerance using combinations of efflux pumps. ACS Synth Biol 4(10):1056–1063. https://doi.org/10.1021/sb500307w
Vancetto GT, Ceccato-Antonini SR (2007) MPK1 gene is required for filamentous growth induced by isoamyl alcohol in Saccharomyces cerevisiae strains from the alcoholic fermentation. Appl Microbiol Biotechnol 75(1):111–115. https://doi.org/10.1007/s00253-006-0795-6
Wang BW, Shi AQ, Tu R, Zhang XL, Wang QH, Bai FW (2012) Branched-chain higher alcohols. Adv Biochem Eng Biotechnol 128:101–118. https://doi.org/10.1007/10_2011_121
Xu P, Qiao K, Ahn WS, Stephanopoulos G (2016) Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.1607295113
Yomano LP, York SW, Ingram LO (1998) Isolation and characterization of ethanol-tolerant mutants of Escherichia coli KO11 for fuel ethanol production. J Ind Microbiol Biotechnol 20(2):132–138
You L, Page L, Feng X, Berla B, Pakrasi HB, Tang YJ (2012) Metabolic pathway confirmation and discovery through (13)C-labeling of proteinogenic amino acids. J Vis Exp 59:e3583. https://doi.org/10.3791/3583
Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, Chen L, Tian W, Tao Y, Kristiansen K, Zhang X, Li S, Yang H, Wang J, Wang J (2010) Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res 20(5):646–654. https://doi.org/10.1101/gr.100677.109
Acknowledgments
We would like to thank Dr. Le You and Dr. Lian He from the Washington University in St. Louis for discussion on 13C-metabolic flux analysis experiments. We also would like to thank Ms. Xianni Qi and Yuanyuan Zhang of Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, for technique assistance in GS-MS analysis.
Funding
This work was supported by the National Basic Research Program (973 Program, 2011CBA00800) and the National Knowledge Innovation Project of the Chinese Academy of Sciences (KSCX2-EW-G-14). QHW was supported by the One Hundred Talent Program of the Chinese Academy of Sciences, and RT was supported by Youth Innovation Promotion Association of Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Wang, B., Guo, Y., Xu, Z. et al. Genomic, transcriptomic, and metabolic characterizations of Escherichia coli adapted to branched-chain higher alcohol tolerance. Appl Microbiol Biotechnol 104, 4171–4184 (2020). https://doi.org/10.1007/s00253-020-10507-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10507-0