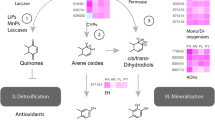
Abstract
The environmental accumulation of polycyclic aromatic hydrocarbons (PAHs) is of great concern due to potential carcinogenic and mutagenic risks, as well as their resistance to remediation. While many fungi have been reported to break down PAHs in environments, the details of gene-based metabolic pathways are not yet comprehensively understood. Specifically, the genome-scale transcriptional responses of fungal PAH degradation have rarely been reported. In this study, we report the genomic and transcriptomic basis of PAH bioremediation by a potent fungal degrader, Dentipellis sp. KUC8613. The genome size of this fungus was 36.71 Mbp long encoding 14,320 putative protein-coding genes. The strain efficiently removed more than 90% of 100 mg/l concentration of PAHs within 10 days. The genomic and transcriptomic analysis of this white rot fungus highlights that the strain primarily utilized non-ligninolytic enzymes to remove various PAHs, rather than typical ligninolytic enzymes known for playing important roles in PAH degradation. PAH removal by non-ligninolytic enzymes was initiated by both different PAH-specific and common upregulation of P450s, followed by downstream PAH-transforming enzymes such as epoxide hydrolases, dehydrogenases, FAD-dependent monooxygenases, dioxygenases, and glycosyl- or glutathione transferases. Among the various PAHs, phenanthrene induced a more dynamic transcriptomic response possibly due to its greater cytotoxicity, leading to highly upregulated genes involved in the translocation of PAHs, a defense system against reactive oxygen species, and ATP synthesis. Our genomic and transcriptomic data provide a foundation of understanding regarding the mycoremediation of PAHs and the application of this strain for polluted environments.






Similar content being viewed by others
References
Abe A, Inoue A, Usami R, Moriya K, Horikoshi K (1995) Degradation of polyaromatic hydrocarbons by organic solvent-tolerant bacteria from deep sea. Biosci Biotechnol Biochem 59(6):1154–1156. https://doi.org/10.1271/bbb.59.1154
Alkio M, Tabuchi TM, Wang XC, Colón-Carmona A (2005) Stress responses to polycyclic aromatic hydrocarbons in Arabidopsis include growth inhibition and hypersensitive response-like symptoms. J Exp Bot 56(421):2983–2994. https://doi.org/10.1093/jxb/eri295
Ben Hamman O, de La Rubia T, Martinez J (1997) Effect of carbon and nitrogen limitation on lignin peroxidase and manganese peroxidase production by Phanerochaete flavido-alba. J Appl Microbiol 83(6):751–757
Cao J, Lai Q, Yuan J, Shao Z (2015) Genomic and metabolic analysis of fluoranthene degradation pathway in Celeribacter indicus P73T. Sci Rep 5:7741. https://doi.org/10.1038/srep07741
Carmona M, Zamarro MT, Blázquez B, Durante-Rodríguez G, Juárez JF, Valderrama JA, Barragán MJ, García JL, Díaz E (2009) Anaerobic catabolism of aromatic compounds: a genetic and genomic view. Microbiol Mol Biol Rev 73(1):71–133. https://doi.org/10.1128/MMBR.00021-08
Casillas RP, Crow SA, Heinze TM, Deck J, Cerniglia CE (1996) Initial oxidative and subsequent conjugative metabolites produced during the metabolism of phenanthrene by fungi. J Ind Microbiol 16(4):205–215. https://doi.org/10.1007/Bf01570023
D'Souza TM, Merrit CS, Reddy CA (1999) Lignin-modifying enzymes of the white rot basidiomycete Ganoderma lucidum. Appl Environ Microbiol 65(12):5307–5313
de Boer W, Folman LB, Summerbell RC, Boddy L (2005) Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29(4):795–811. https://doi.org/10.1016/j.femsre.2004.11.005
Deshmukh R, Khardenavis AA, Purohit HJ (2016) Diverse metabolic capacities of fungi for bioremediation. Indian J Microbiol 56(3):247–264. https://doi.org/10.1007/s12088-016-0584-6
Field JA, de Jong E, Feijoo Costa G, de Bont JA (1992) Biodegradation of polycyclic aromatic hydrocarbons by new isolates of white rot fungi. Appl Environ Microbiol 58(7):2219–2226
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Report 13(3):207–209. https://doi.org/10.1007/Bf02670897
Gan S, Lau EV, Ng HK (2009) Remediation of soils contaminated with polycyclic aromatic hydrocarbons (PAHs). J Hazard Mater 172(2–3):532–549. https://doi.org/10.1016/j.jhazmat.2009.07.118
Gao R, Hao DC, Hu WL, Song S, Li SY, Ge GB (2019) Transcriptome profile of polycyclic aromatic hydrocarbon-degrading fungi isolated from Taxus rhizosphere. Curr Sci India 116(7):1218–1228. https://doi.org/10.18520/cs/v116/i7/1218-1228
Ghosal D, Ghosh S, Dutta TK, Ahn Y (2016) Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): a review. Front Microbiol 7:1369. https://doi.org/10.3389/fmicb.2016.01369
Gnerre S, MacCallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, Berlin AM, Aird D, Costello M, Daza R, Williams L, Nicol R, Gnirke A, Nusbaum C, Lander ES, Jaffe DB (2011) High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA 108(4):1513–1518. https://doi.org/10.1073/pnas.1017351108
Gran-Scheuch A, Fuentes E, Bravo DM, Jiménez JC, Pérez-Donoso JM (2017) Isolation and characterization of phenanthrene degrading bacteria from diesel fuel-contaminated Antarctic soils. Front Microbiol 8:1634. https://doi.org/10.3389/fmicb.2017.01634
Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I (2014) MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42(Database issue):D699–D704. https://doi.org/10.1093/nar/gkt1183
Habe H, Omori T (2003) Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci Biotechnol Biochem 67(2):225–243. https://doi.org/10.1271/bbb.67.225
Hadibarata T, Tachibana S, Itoh K (2007) Biodegradation of phenanthrene by fungi screened from nature. Pak J Biol Sci 10(15):2535–2543
Harms H, Schlosser D, Wick LY (2011) Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat Rev Microbiol 9(3):177–192. https://doi.org/10.1038/nrmicro2519
Jeong CB, Kim DH, Kang HM, Lee YH, Kim HS, Kim IC, Lee JS (2017) Genome-wide identification of ATP-binding cassette (ABC) transporters and their roles in response to polycyclic aromatic hydrocarbons (PAHs) in the copepod Paracyclopina nana. Aquat Toxicol 183:144–155. https://doi.org/10.1016/j.aquatox.2016.12.022
Johansson I, van Bavel B (2003) Levels and patterns of polycyclic aromatic hydrocarbons in incineration ashes. Sci Total Environ 311(1–3):221–231. https://doi.org/10.1016/S0048-9697(03)00168-2
Kadri T, Rouissi T, Kaur Brar S, Cledon M, Sarma S, Verma M (2017) Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by fungal enzymes: a review. J Environ Sci (China) 51:52–74. https://doi.org/10.1016/j.jes.2016.08.023
Lee H, Jang Y, Choi YS, Kim MJ, Lee J, Lee H, Hong JH, Lee YM, Kim GH, Kim JJ (2014) Biotechnological procedures to select white rot fungi for the degradation of PAHs. J Microbiol Methods 97:56–62. https://doi.org/10.1016/j.mimet.2013.12.007
Lewis DFV (2003) Human cytochromes P450 associated with the phase 1 metabolism of drugs and other xenobioties: a compilation of substrates and inhibitors of the CYP1, CYP2 and CYP3 families. Curr Med Chem 10(19):1955–1972. https://doi.org/10.2174/0929867033456855
Mao J, Guan WW (2016) Fungal degradation of polycyclic aromatic hydrocarbons (PAHs) by Scopulariopsis brevicaulis and its application in bioremediation of PAH-contaminated soil. Acta Agr Scand B-S P 66(5):399–405. https://doi.org/10.1080/09064710.2015.1137629
Marco-Urrea E, García-Romera I, Aranda E (2015) Potential of non-ligninolytic fungi in bioremediation of chlorinated and polycyclic aromatic hydrocarbons. New Biotechnol 32(6):620–628. https://doi.org/10.1016/j.nbt.2015.01.005
Mastrangelo G, Fadda E, Marzia V (1996) Polycyclic aromatic hydrocarbons and cancer in man. Environ Health Perspect 104(11):1166–1170. https://doi.org/10.2307/3432909
Min B, Grigoriev IV, Choi IG (2017) FunGAP: fungal genome annotation pipeline using evidence-based gene model evaluation. Bioinformatics 33(18):2936–2937. https://doi.org/10.1093/bioinformatics/btx353
Mulligan CN (2005) Environmental applications for biosurfactants. Environ Pollut 133(2):183–198. https://doi.org/10.1016/j.envpol.2004.06.009
Peng RH, Xiong AS, Xue Y, Fu XY, Gao F, Zhao W, Tian YS, Yao QH (2008) Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol Rev 32(6):927–955. https://doi.org/10.1111/j.1574-6976.2008.00127.x
Pozdnyakova NN (2012) Involvement of the ligninolytic system of white-rot and litter-decomposing fungi in the degradation of polycyclic aromatic hydrocarbons. Biotechnol Res Int 2012:243217–243220. https://doi.org/10.1155/2012/243217
Ron EZ, Rosenberg E (2001) Natural roles of biosurfactants. Environ Microbiol 3(4):229–236
Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, Levasseur A, Lombard V, Morin E, Otillar R, Lindquist EA, Sun H, LaButti KM, Schmutz J, Jabbour D, Luo H, Baker SE, Pisabarro AG, Walton JD, Blanchette RA, Henrissat B, Martin F, Cullen D, Hibbett DS, Grigoriev IV (2014) Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci U S A 111(27):9923–9928. https://doi.org/10.1073/pnas.1400592111
Sena HH, Sanches MA, Rocha DFS, Segundo W, de Souza ES, de Souza JVB (2018) Production of biosurfactants by soil fungi isolated from the Amazon forest. Int J Microbiol 2018:5684261–5684268. https://doi.org/10.1155/2018/5684261
Schützendübel A, Majcherczyk A, Johannes C, Hüttermann A (1999) Degradation of fluorene, anthracene, phenanthrene, fluoranthene, and pyrene lacks connection to the production of extracellular enzymes by Pleurotus ostreatus and Bjerkandera adusta. Int Biodeterior Biodegradation 43(3):93–100. https://doi.org/10.1016/S0964-8305(99)00035-9
Syed K, Doddapaneni H, Subramanian V, Lam YW, Yadav JS (2010) Genome-to-function characterization of novel fungal P450 monooxygenases oxidizing polycyclic aromatic hydrocarbons (PAHs). Biochem Bioph Res Co 399(4):492–497. https://doi.org/10.1016/j.bbrc.2010.07.094
Yalkowsky SH, He Y, Jain P (2004) Handbook of aqueous solubility data. Choice 41(5):941
Yu GC, Wang LG, Han YY, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
Acknowledgments
This work is supported by the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01337602) Rural Development Administration and New and Renewable Energy Core Technology Program of the Korea Institute of Energy Technology Evaluation and Planning (KETEP) grants from the Ministry of Trade, Industry and Energy (No. 20173010092460), Republic of Korea. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Park, H., Min, B., Jang, Y. et al. Comprehensive genomic and transcriptomic analysis of polycyclic aromatic hydrocarbon degradation by a mycoremediation fungus, Dentipellis sp. KUC8613. Appl Microbiol Biotechnol 103, 8145–8155 (2019). https://doi.org/10.1007/s00253-019-10089-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10089-6