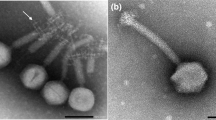
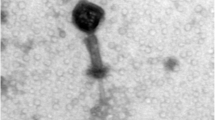
Abstract
Following the emergence of antibiotic-resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant Staphylococcus pseudintermedius (MRSP), phage therapy has attracted significant attention as an alternative to antibiotic treatment. Bacteriophages belonging to kayvirus (previously known as Twort-like phages) have broad host range and are strictly lytic in Staphylococcus spp. Previous work revealed that kayvirus ɸSA039 has a host-recognition mechanism distinct from those of other known kayviruses: most of kayviruses use the backbone of wall teichoic acid (WTA) as their receptor; by contrast, ɸSA039 uses the β-N-acetylglucosamine (β-GlcNAc) residue in WTA. In this study, we found that ɸSA039 could switch its receptor to be able to infect S. aureus lacking the β-GlcNAc residue by acquiring a spontaneous mutation in open reading frame (ORF) 100 and ORF102. Moreover, ɸSA039 could infect S. pseudintermedius, which has a different WTA structure than S. aureus. By comparison, with newly isolated S. pseudintermedius–specific phage (SP phages), we determined that glycosylation in WTA of S. pseudintermedius is essential for adsorption of SP phages, but not ɸSA039. Finally, we describe a novel strategy of S. aureus which protects the bacteria from infection of SP phages. Notably, glycosylation of ribitol phosphate (RboP) WTA by TarM or/and TarS prevents infection of S. aureus by SP phages. These findings could help to establish a new strategy for the treatment of S. aureus and S. pseudintermedius infection, as well as provide valuable insights into the biology of phage–host interactions.




Similar content being viewed by others
References
Alves DR, Gaudion A, Bean JE, Perez Esteban P, Arnot TC, Harper DR, Kot W, Hansen LH, Enright MC, Jenkins ATA (2014) Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl Environ Microbiol 80:6694–6703. https://doi.org/10.1128/AEM.01789-14
Asheshov EA, Jevons MP (1963) The effect of heat on the ability of a host strain to support the growth of a Staphylococcusphage. J Gen Microbiol 31:97–107. https://doi.org/10.1099/00221287-31-1-97
Azam AH, Tanji Y (2019a) Bacteriophage-host arm race : An update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl Microbiol Biotechnol 103:2121–2131. https://doi.org/10.1007/s00253-019-09629-x
Azam AH, Tanji Y (2019b) Peculiarities of Staphylococcus aureus phages and their possible application in phage therapy. Appl Microbiol Biotechnol 103(11):4279–4289. https://doi.org/10.1007/s00253-019-09810-2
Azam AH, Hoshiga F, Takeuchi I, Miyanaga K, Tanji Y (2018) Analysis of phage resistance in Staphylococcus aureus SA003 reveals different binding mechanisms for the closely related Twort-like phages ϕSA012 and ϕSA039. Appl Microbiol Biotechnol 102:8963–8977
Bannoehr J, Ben Zakour NL, Waller AS, Guardabassi L, Thoday KL, Van Den Broek AHM, Fitzgerald JR (2007) Population genetic structure of the Staphylococcus intermedius group: Insights into agrdiversification and the emergence of methicillin-resistant strains. J Bacteriol 189:8685–8692. https://doi.org/10.1128/JB.01150-07
Bannoehr J, Franco A, Iurescia M, Battisti A, Fitzgerald JR (2009) Molecular diagnostic identification of Staphylococcus pseudintermedius. J Clin Microbiol 47:469–471. https://doi.org/10.1128/JCM.01915-08
Blaiotta G, Fusco V, Ercolini D, Pepe O, Coppola S (2010) Diversity of Staphylococcus species strains based on partial kat (catalase) gene sequences and design of a PCR-restriction fragment length polymorphism assay for identification and differentiation of coagulase-positive species (S. aureus, S. delphini, S. hyicus, S. intermedius, S. pseudintermedius, and S. schleiferi subsp. coagulans). J Clin Microbiol 48:192–201. https://doi.org/10.1128/JCM.00542-09
Brown S, Xia G, Luhachack LG, Campbell J, Meredith TC, Chen C, Winstel V, Gekeler C, Irazoqui JE, Peschel A, Walker S (2012) Methicillin resistance in Staphylococcus aureusrequires glycosylated wall teichoic acids. Proc Natl Acad Sci 109:18909–18914. https://doi.org/10.1073/pnas.1209126109
Brown S, Santa Maria JP, Walker S (2013) Wall teichoic acids of Gram-positive bacteria. Annu Rev Microbiol 67:313–336. https://doi.org/10.1146/annurev-micro-092412-155620
Brussow H, Canchaya C, Hardt W-D (2004) Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602. https://doi.org/10.1128/MMBR.68.3.560-602.2004
Capparelli R, Nocerino N, Lanzetta R, Silipo A, Amoresano A, Giangrande C, Becker K, Blaiotta G, Evidente A, Cimmino A, Iannaccone M, Parlato M, Medaglia C, Roperto S, Roperto F, Ramunno L, Iannelli D (2010) Bacteriophage-resistant Staphylococcus aureus mutant confers broad immunity against staphylococcal infection in mice. PLoS One 5. https://doi.org/10.1371/journal.pone.0011720
Chen W, Zhang Y, Yeo WS, Bae T, Ji Q (2017) Rapid and efficient genome editing in Staphylococcus aureus by using an engineered CRISPR/Cas9 system. J Am Chem Soc 139:3790–3795. https://doi.org/10.1021/jacs.6b13317
Cui Z, Song Z, Wang Y, Zeng L, Shen W, Wang Z, Li Q, He P, Qin J, Guo X (2012) Complete genome sequence of wide-host-range Staphylococcus aureus phage JD007. J Virol 86:13880–13881. https://doi.org/10.1128/JVI.02728-12
Cui Z, Guo X, Dong K, Zhang Y, Li Q, Zhu Y, Zeng L, Tang R, Li L (2017) Safety assessment of Staphylococcus phages of the family Myoviridae based on complete genome sequences. Sci Rep 7. https://doi.org/10.1038/srep41259
Deghorain M, Bobay LM, Smeesters PR, Bousbata S, Vermeersch M, Perez-Morga D, Drèze PA, Rocha EPC, Touchon M, Van Melderen L (2012) Characterization of novel phages isolated in coagulase-negative staphylococci reveals evolutionary relationships with Staphylococcus aureusphages. J Bacteriol 194:5829–5839. https://doi.org/10.1128/JB.01085-12
Endl J, Seidl HP, Fiedler F, Schleifer KH (1983) Chemical composition and structure of cell wall teichoic acids of staphylococci. Arch Microbiol 135:215–223. https://doi.org/10.1007/BF00414483
Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG, Walsh CT (2002) The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci 99:7687–7692
Gutiérrez D, Fernández L, Rodríguez A, García P (2018) Practical method for isolation of phage deletion mutants. Methods Protoc 1:6. https://doi.org/10.3390/mps1010006
Hyman P, Abedon ST (2010) Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70:217–248
Iwano H, Inoue Y, Takasago T, Kobayashi H, Furusawa T, Taniguchi K, Fujiki J, Yokota H, Usui M, Tanji Y, Hagiwara K, Higuchi H, Tamura Y (2018) Bacteriophage ΦSA012 has a broad host range against Staphylococcus aureus and effective lytic capacity in a mouse mastitis model. Biology (Basel) 7:8. https://doi.org/10.3390/biology7010008
Jeong DW, Cho H, Lee H, Li C, Garza J, Fried M, Bae T (2011) Identification of the P3 promoter and distinct roles of the two promoters of the SaeRS two-component system in Staphylococcus aureus. J Bacteriol 193:4672–4684. https://doi.org/10.1128/JB.00353-11
Kloos WE, Bannerman TL (1994) Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev 7:117–140
Kurokawa K, Jung DJ, An JH, Fuchs K, Jeon YJ, Kim NH, Li X, Tateishi K, Park JA, Xia G, Matsushita M, Takahashi K, Park HJ, Peschel A, Lee BL (2013) Glycoepitopes of staphylococcal wall teichoic acid govern complement-mediated opsonophagocytosis via human serum antibody and mannose-binding lectin. J Biol Chem 288:30956–30968. https://doi.org/10.1074/jbc.M113.509893
Li X, Gerlach D, Du X, Larsen J, Stegger M, Kuhner P, Peschel A, Xia G, Winstel V (2015) An accessory wall teichoic acid glycosyltransferase protects Staphylococcus aureus from the lytic activity of Podoviridae. Sci Rep 5:17219. https://doi.org/10.1038/srep17219
Li X, Koç C, Kühner P, Stierhof Y-D, Krismer B, Enright MC, Penadés JR, Wolz C, Stehle T, Cambillau C, Peschel A, Xia G (2016) An essential role for the baseplate protein Gp45 in phage adsorption to Staphylococcus aureus. Nat Publ Group 6:26455. https://doi.org/10.1038/srep26455
Łobocka M, Hejnowicz MS, Dabrowski K, Gozdek A, Kosakowski J, Witkowska M, Ulatowska MI, Weber-Dabrowska B, Kwiatek M, Parasion S, Gawor J, Kosowska H, Głowacka A (2012) Genomics of Staphylococcal twort-like phages - potential therapeutics of the post-antibiotic era. Adv Virus Res 83:143–216. https://doi.org/10.1016/B978-0-12-394438-2.00005-0
Moodley A, Stegger M, Ben Zakour NL, Fitzgerald JR, Guardabassi L (2009) Tandem repeat sequence analysis of staphylococcal protein A (spa) gene in methicillin-resistant Staphylococcus pseudintermedius. Vet Microbiol 135:320–326. https://doi.org/10.1016/j.vetmic.2008.09.070
Morita M, Fischer CR, Mizoguchi K, Yoichi M, Oda M, Tanji Y, Unno H (2002) Amino acid alterations in Gp38 of host range mutants of PP01 and evidence for their infection of an OmpC null mutant of Escherichia coli O157:H7. FEMS Microbiol Lett 216:243–248. https://doi.org/10.1016/S0378-1097(02)01025-X
O’Flaherty S, Coffey A, Edwards R, Meaney W, Fitzgerald GF, Ross RP (2004) Genome of staphylococcal phage K: A new lineage of myoviridae infecting Gram-positive bacteria with a low G+C content. J Bacteriol 186:2862–2871. https://doi.org/10.1128/JB.186.9.2862-2871.2004
Osada K, Takeuchi I, Miyanaga K, Tanji Y (2017) Coevolution between Staphylococcus aureus isolated from mastitic milk and its lytic bacteriophage φSA012 in batch co-culture with serial transfer. Biochem Eng J 126:16–23. https://doi.org/10.1016/j.bej.2017.06.022
Pompilio A, De Nicola S, Crocetta V, Guarnieri S, Savini V, Carretto E, Di Bonaventura G (2015) New insights in Staphylococcus pseudintermedius pathogenicity: Antibiotic-resistant biofilm formation by a human wound-associated strain. BMC Microbiol 15. https://doi.org/10.1186/s12866-015-0449-x
Sakoulas G, Eliopoulos GM, Fowler VG, Moellering RC, Novick RP, Lucindo N, Yeaman MR, Bayer AS (2005) Reduced susceptibility of Staphylococcus aureus to vancomycin and platelet microbicidal protein correlates with defective autolysis and loss of accessory gene regulator (agr) function. Antimicrob Agents Chemother 49:2687–2692. https://doi.org/10.1128/AAC.49.7.2687-2692.2005
Synnott AJ, Kuang Y, Kurimoto M, Yamamichi K, Iwano H, Tanji Y (2009) Isolation from sewage influent and characterization of novel Staphylococcus aureus bacteriophages with wide host ranges and potent lytic capabilities. Appl Environ Microbiol 75:4483–4490. https://doi.org/10.1128/AEM.02641-08
Takeuchi I, Osada K, Azam AH, Asakawa H, Miyanaga K, Tanji Y (2016) The presence of two receptor-binding proteins contributes to the wide host range of staphylococcal twort-like phages. Appl Environ Microbiol 82:5763–5774. https://doi.org/10.1128/AEM.01385-16
Weidenmaier C, Peschel A, Xiong Y, Kristian SA, Dietz K, Yeaman MR, Bayer AS (2005) Lack of wall teichoic acids in Staphylococcus aureus leads to reduced interactions with endothelial cells and to attenuated virulence in a rabbit model of endocarditis. J Infect Dis 191:1771–1777. https://doi.org/10.1086/429692
Winstel V, Liang C, Sanchez-Carballo P, Steglich M, Munar M, Broker BM, Penadés JR, Nübel U, Holst O, Dandekar T, Peschel A, Xia G (2013) Wall teichoic acid structure governs horizontal gene transfer between major bacterial pathogens. Nat Commun 4:2345. https://doi.org/10.1038/ncomms3345
Winstel V, Sanchez-Carballo P, Holst O, Xia G, Peschel A (2014) Biosynthesis of the unique wall teichoic acid of Staphylococcus aureus lineage ST395. MBio 5(2):e00869. https://doi.org/10.1128/mBio.00869-14
Xia G, Corrigan RM, Winstel V, Goerke C, Gründling A, Peschel A (2011) Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J Bacteriol 193:4006–4009. https://doi.org/10.1128/JB.01412-10
Xia G, Maier L, Sanchez-Carballo P, Li M, Otto M, Holst O, Peschel A (2010) Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J Biol Chem 285:13405–13415. https://doi.org/10.1074/jbc.M109.096172
Xia G, Wolz C (2014) Phages of Staphylococcus aureus and their impact on host evolution. Infect Genet Evol 21:593–601. https://doi.org/10.1016/j.meegid.2013.04.022
Zhu X, Liu D, Singh AK, Drolia R, Bai X, Tenguria S, Bhunia AK (2018) Tunicamycin mediated inhibition of wall teichoic acid affects Staphylococcus aureus and Listeria monocytogenes cell morphology, biofilm formation and virulence. Front Microbiol 9:1352. https://doi.org/10.3389/fmicb.2018.01352
Acknowledgments
We thank Professor Kenji Kurokawa at the Faculty of Pharmaceutical Science of Nagasaki International University for providing us with the deletion mutant of RN4220.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animal performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 385 kb)
Rights and permissions
About this article
Cite this article
Azam, A.H., Kadoi, K., Miyanaga, K. et al. Analysis host-recognition mechanism of staphylococcal kayvirus ɸSA039 reveals a novel strategy that protects Staphylococcus aureus against infection by Staphylococcus pseudintermedius Siphoviridae phages. Appl Microbiol Biotechnol 103, 6809–6823 (2019). https://doi.org/10.1007/s00253-019-09940-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09940-7