Abstract
The microbial degradation of plant-derived compounds under salinity stress remains largely underexplored. The pretreatment of lignocellulose material, which is often needed to improve the production of lignocellulose monomers, leads to high salt levels, generating a saline environment that raises technical considerations that influence subsequent downstream processes. Here, we constructed halotolerant lignocellulose degrading microbial consortia by enriching a salt marsh soil microbiome on a recalcitrant carbon and energy source, i.e., wheat straw. The consortia were obtained after six cycles of growth on fresh substrate (adaptation phase), which was followed by four cycles on pre-digested (highly-recalcitrant) substrate (stabilization phase). The data indicated that typical salt-tolerant bacteria made up a large part of the selected consortia. These were “trained” to progressively perform better on fresh substrate, but a shift was observed when highly recalcitrant substrate was used. The most dominant bacteria in the consortia were Joostella marina, Flavobacterium beibuense, Algoriphagus ratkowskyi, Pseudomonas putida, and Halomonas meridiana. Interestingly, fungi were sparsely present and negatively affected by the change in the substrate composition. Sarocladium strictum was the single fungal strain recovered at the end of the adaptation phase, whereas it was deselected by the presence of recalcitrant substrate. Consortia selected in the latter substrate presented higher cellulose and lignin degradation than consortia selected on fresh substrate, indicating a specialization in transforming the recalcitrant regions of the substrate. Moreover, our results indicate that bacteria have a prime role in the degradation of recalcitrant lignocellulose under saline conditions, as compared to fungi. The final consortia constitute an interesting source of lignocellulolytic haloenzymes that can be used to increase the efficiency of the degradation process, while decreasing the associated costs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lignocellulosic plant biomass is the most abundant global carbon source. Aside its availability and low cost, its utilization can attenuate the conflict between food and energy crops (Kinet et al. 2015). However, the main obstacle in its widespread application is the high cost of the pretreatments, which are necessary to open the intricate polysaccharide structure. Such pretreatments enhance the accessibility of enzymatic attack (Talebnia et al. 2010) and decrease the proportion of crystalline cellulose and lignin content, the two main causes of the recalcitrance of lignocellulose. Overcoming this recalcitrance is fundamental for getting access to the polymers that yield sugar monomers, which can be transformed in valuable compounds such as sustainable biomaterials, biofuel, and biochemicals (Khoo et al. 2016).
In the past years, three different pretreatment processes have been proposed to improve the digestibility of lignocellulose materials. These aimed to foster (1) the degradation of hemicellulose, by acid or hot water treatment, (2) that of lignin, by alkaline pretreatment to break the lignin-carbohydrate linkage bond, and (3) the generic disruption of the matrix by thermal treatment (Brethauer and Studer 2015). Such pretreatments not only increase the global cost of the bioprocess but also generate diverse compounds that interfere with downstream processes (Jönsson and Martin 2016; Rabemanolontsoa and Saka 2016).
A promising new pretreatment method is based on the application of ionic liquids (ILs), organic salts (“green solvents”) (Sun et al. 2016) that are liquid at room temperature. Using ILs, lignocellulose biomass is exposed to highly saline conditions that disrupt the rigid lignocellulose structure, leading to a considerable reduction in cristallinity and increased accessibility to enzymatic attack. However, when using acid/base treatment or ILs, subsequent enzymatic hydrolysis of the substrate can only be performed after several washing steps aiming at salt removal, as salt often inhibits enzymatic activity. The use of haloenzymes (or enzymes tolerant to high salinity) (Gunny et al. 2014) could represent a sound alternative strategy to increase the efficiency and reduce the cost of the bioprocess.
Dilution-to-stimulation has been used as a successful method to enrich microbial consortia capable of degrading plant biomass and their respective enzymes (Brossi et al. 2015; Maruthamuthu et al. 2016). These consortia have been obtained from a variety of sources (Cortes-Tolalpa et al. 2016) and are often capable of degrading a range of lignocellulose materials (Okeke and Lu 2011; Brossi et al. 2015). For instance, we have shown that consortia obtained from different microbial sources naturally enriched in lignocellulose material quickly reach a stabilization phase (phase of relative stability of the consortium in terms of composition and activity) during the enrichment process (Cortes-Tolalpa et al. 2016). Although the various consortia did not differ in their final degradation potential, they reached this through different activities, as they differed in their enzymatic pools. Thus, the source of the inoculum used for the enrichment clearly influenced the final outcome and type of process. Despite the success of this approach, which leads to consortia capable of “attacking” or consuming the most labile part of the substrate, these consortia have been obtained under “low” salt concentrations. Given the importance of the microbial source, the development of such consortia using halotolerant microbes could provide an interesting perspective.
The aim of this study was to examine whether it is possible to obtain a halotolerant microbial consortium capable of degrading lignocellulose biomass (raw wheat straw) at high rate under high-salt conditions. For that, we used as inoculum the microbial community obtained from salt marsh soil from a the island of Schiermonnikoog, the Netherlands. This was previously found to be adapted to high-salt concentrations and to harbor key genes involved in lignocellulose degradation (Dini-Andreote et al. 2014; Wang et al. 2016). In addition, to generate consortia with high degradation potential under high-salt conditions, selection on pre-digested recalcitrant substrate was applied.
Methods
Culture media and lignocellulose substrate
For the experiment, we used a the mineral medium solution MMS (7 g/L Na2HPO4·2H2O; 2 g/L K2HPO4; 1 g/L (NH4)2SO4; 0.1 g/L Ca (NO3)2·4H2O; 0.2 g/L MgCl2·6H2O g/L, pH 7.2) (Cortes-Tolalpa et al. 2016), supplemented with 25 g per liter of NaCl. The medium was further supplemented with vitamin solution (0.1 g Ca-pantothenate, 0.1 g cyanocobalamine, 0.1 g nicotinic acid, 0.1 g pyridoxal, 0.1 g riboflavin, 0.1 g thiamin, 0.01 g biotin, 0.1 g folic acid; H2O 1 L) and trace metal solution (2.5 g/L EDTA; 1.5 g/L FeSO4; 0.025 g/L CoCl2; 0.025 g/L ZnSO4; 0.015 g/L MnCl2; 0.015 g/L NaMoO4; 0.01 g/L NiCl2; 0.02 g/L H3BO3; 0.005 g/L CuCl2). “Raw wheat straw” used as lignocellulose source, was air-dried (50 °C) before cutting it into pieces of about 5 cm length and then the pieces were thoroughly ground, using a mill hammer, to pieces ≤ 1 mm. No pre-treatment was performed (untreated raw substrate). Sterility of the substrate was verified following plating on trypticase soy agar (TSA) plates. All chemicals and reagents used in this work were of analytic molecular biology grade (Sigma-Aldrich, Darmstadt, Germany).
Sample collection
The source of the microbial community used in this experiment was soil from Schiermonnikoog island (53°29’ N 6°10′ E), 10-g of surface soil (0–10 cm) representative of the 105-year old plot located at the end of the natural primary succession observed in this island (Wang et al. 2016), the soil samples were thoroughly mixed. These soils are characterized by pH varying from 7.4–7.6 and sodium concentration from 3541 ± 170 to 5188 ± 624 mg dm−3, depending on the period of the year (Dini-Andreote et al. 2014). Cell suspension was prepared by adding 10 g of the soil to 250 mL flasks containing 10 g of sterile gravel in 90 mL of MMS. The suspension was shaken for 30 min at 200 rpm (room temperature).
Enriched consortia
To start the enrichment, 250 μL of the suspension was added to each of triplicate 100-ml Erlenmeyer flasks containing 25 mL of MMS supplemented with 1% (w/v) sterilized wheat straw, 25 μL of vitamin and 25 μl of trace metal solution. Flasks were incubated at 28 °C, with shaking at 180 rpm. Cultures were monitored by counting cells in a Bürker-Türk chamber every day. Experiments started with around 5 log cells/mL. Once the systems had reached around 9 log cells/mL (and straw had visually been degraded), 25 μL of culture was transferred to 25 mL of fresh medium (dilution 10−3). During the first part of the enrichment, from transfer one to six—the adaptation phase—we used fresh wheat straw. In the second part of the experiment, from transfer seven to ten—the stabilization phase—we used recalcitrant wheat straw. This consisted of the sterilized substrate recovered at the end of the adaptation phase (transfers five and six), partially consumed by microbial consortia, and therefore encompassing only the most recalcitrant structure of the substrate (Supplemental Fig. S1). Following each transfer (T), part of the bred consortia was stored in 20% glycerol at − 80 °C. The consortia of the T1, T3, T6, T7, and T10 flasks were used for all subsequent analyses, as detailed below. As controls, we used microbial sources in MMS without substrate (CA 1, 2, 3) as well as MMS plus substrate without inoculum (CB 1, 2, 3). Before starting the enrichment Erlenmeyer flasks containing 25 mL lignocellulose, media were autoclaved at 121 °C for 27 min.
DNA extraction
One mL of selected cultures was used for community DNA extraction using the “Power Soil” DNA extraction kit (inoculum source) (MoBio® Laboratories Inc., Carlsbad, USA) and the UltraClean DNA Isolation Kit (each enriched consortium and isolates). The instructions of the manufacturer were followed, except that the resuspension of the DNA from the inoculum sources was in 60 μL resuspension fluid.
PCR followed by denaturing gradient gel electrophoresis (PCR-DGGE)
Total community DNA was used as the template for amplification of the partial 16S rRNA gene fragment by PCR with primers F968 with a GC clamp attached to the 5′-end and universal bacterial primer R1401.1b. For ITS1 amplification, primers EF4/ITS4 were used; this PCR was followed by a second amplification with primers ITS1f-GCITS2. Primer sequences, the reaction mixtures, and cycling conditions have been described (Brons and van Elsas 2008; Pereira e Silva et al. 2012). The DGGE was performed as reported by Cortes-Tolalpa et al. (2016). The DGGE patterns were then transformed to a band-matching table using GelCompar II software (Applied Maths, Sint Martens Latem, Belgium).
Quantitative PCR (q-PCR)
The 16S rRNA gene region V5-V6 (bacteria), as well as the ITS1 region (fungi), were amplified using 1 ng of community DNA as the template and primers 16SFP/16SRP and 5.8S/ITS1 (Pereira e Silva et al. 2012), respectively. Standard curves were constructed using serial dilutions of cloned 16S rRNA gene and ITS1 fragments from Serratia plymuthica (KF495530) and Coniochaeta ligniaria (KF285995), respectively. The gene target quantification was performed, in triplicate, in an ABI Prism 7300 Cycler (Applied Biosystem, Lohne, Germany).
Bacterial community sequencing and analyses
Amplicons of 250 bp were generated based on primers amplifying the V4-V5 of the 16S rRNA gene. PCR amplifications were conducted in triplicate reactions for each of the 18 samples with the 515F/806R primer set (Supplemental Table S1). PCR and sequencing were performed using a standard protocol (Caporaso et al. 2012). Illumina MiSeq sequencing was performed at GENEWIZ (South Plainfield, USA). We processed the raw data using the “quantitative insight into microbial ecology” (QIIME) software, version 1.91. The sequences were de-multiplexed and quality-filtered using split_libraries_fastq.py default parameters (Bokulich et al. 2013). The derived sequences were then clustered into operational taxonomic units (OTUs) using open-reference OTU picking against the Greengenes reference OTU data base with a 97% similarity threshold (Rideout et al. 2014). Then, we performed quality–filtering to discard OTUs present at very low abundance (< 0.005%) of the total number of sequences (Bokulich et al. 2013). An even sampling depth of 20,000 sequences per sample was used for assessing α- and β-diversity measures. Metrics for α-diversity were Chao1 index (estimated species richness) and Shannon index (quantitative measure of species). β-diversity analyses among the final consortia were performed using unweighted UniFrac distance matrix. Matrix similarity, PERMANOVA, and principal coordinate analyses (PCA), were performed by using phyloseq (McMurdie and Holmes 2013). Differential OTU abundance was calculated using DESeq2 with phyloseq (Supplemental Fig. S2) (Love et al. 2014; Mcmurdie et al. 2014). The comparison was made between sequential transfers (inocula-T1, T1-T3, T3-T6, T6-T7, T7-T10) and between the two main phases, adaption and stabilization phase, respectively.
Isolation and identification of bacterial and fungi
From transfers 6 and 10, we isolated bacterial and fungal strains, using R2A (BD Difco®, Detroit, USA) and potato dextrose agar (PDA) (Duchefa Biochemie BV, Haarlem, The Netherlands), respectively. The isolation part can be found in Electronic supplemental material 1 (ESM 1). The primer pair U1406R and B8F was used for amplification of the 16S rRNA gene of bacterial strains, in the following PCR: initial denaturation at 95 °C for 5 min; 35 cycles of 95 °C for 1 min, 52 °C for 30 s, 72 °C for 2 min and final extension at 72 °C for 7 min. For identification of fungal strains the primers EF4 and ITS4 were used for amplification of the ITS1 region of the 18S rRNA gene, according to the following PCR : initial denaturation at 95 °C for 5 min; 34 cycles of 94 °C for 30 s, 55 °C for 30 s 72 °C for 1 min 30 s and final extension at 72 °C for 5 min. The amplicons were sequenced by Sanger technology (LGC Genomics, Lückenwalde, Germany) and the sequence of the PCR product was further used for bacterial and fungal identification. Taxonomic assignments of the sequences were done using BLAST-N (http://blast.stva.ncbi.nlm.nih.gov/Blast.cgi). We used the best BLAST hit affiliation for taxonomic assignment with a cutoff of 97 and 95% of identity of bacteria and fungi, respectively, and 95% of coverage. Sequences are publicly available in the GenBank database under accession numbers MF619963 to MF620009 (Tables 3 and 4). The recovered strains have been deposited in the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany).
Matching bacterial strains with abundant OTUs
The recovered bacterial strains were linked to the OTUs based on sequence similarity. The almost-full-length 16S rRNA gene sequences from the strains were compared—in the specific V4-V5 region—to the sequences of the abundant OTUs using ClustalW. Phylogenetic analyses (pairwise distance) were conducted with MEGA v6 (Tamura et al. 2013) using Maximum Likelihood evolutionary distances that were computed using the Kimura-2 parameter method. The branch node strengths were tested with bootstrap analyses (1000 replications).
Screening of lignocellulolytic enzyme production in recovered bacterial strains
Cellulases and hemicellulases in bacterial strains were detected by model substrate coupled to chromogenic compounds. The compounds 5-bromo-4-chloro-3-indolyl α-D-glucopyranoside (X-glu), 5-bromo-4-chloro-3-indolyl β-D-cellobioside (X-cell), 5-bromo-4-chloro-3-indolyl α-D-mannopyranoside (X-man), 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal), 5-bromo-4-chloro-3-indolyl β-D-xylopyranoside (X-xyl), and 5-bromo-4-chloro-3-indolyl β-fucopyranoside (X-fuc) (Sigma-Aldrich, Darmstadt, Germany) were used to detect the production and activity of α-glucosidase, cellobiohydrolases, α-mannosidase, β-galactosidase, β-xylosidase, and α-fucosidase enzymatic activity, respectively (Cortes-Tolalpa et al. 2016). The strains were spread in duplicate on R2A plates containing 1 M NaCl and each one of the chromogenic compounds listed above. The plates were incubated for 48 h at 28 °C. A positive enzymatic activity was observed as a blue colony growing on the plate.
Lignocellulose degradation by selected halotolerant consortia
The final microbial consortia from transfers 1, 3, 6, 7, and 10 were incubated with 1% (w/v) mulched wheat straw under the culture condition that was previously described. After incubation, the final remaining particulate wheat straw was recovered from the microcosm flasks; the substrate was washed to remove microbial cells and sieved to obtain the degraded particles. The degradation rates of the components of the substrate, before and after incubation, were determined by Fourier-transformed infrared (FTIR) spectra (Adapa et al. 2011; Xu et al. 2013). All FTIR measurements were carried out on oven-dried material (50 °C, 24 h). Thirty-two scans were run per sample; all spectra between 800 and 1800 cm1 were used for the analyse (Krasznai et al. 2012). Each sample (calibration and consortium samples) was analyzed in triplicate. All spectra were subjected to baseline correction and then corrected for physical effects by second derivative Savitzky-Golay treatment (FitzPatrick et al. 2012). Correction and analysis using partial least squares (PLS) regression were conducted using Unscrambler X v.10 (CAMO, Woodbridge, USA). A mathematical model was created on the basis of a calibration with standard mixtures, consisting of hemicellulose (proxy beechwood xylan, ≥ 90%, Sigma-Aldrich, Steinheim, Germany), cellulose (powder, D-516, Macherey-Nagel, Düren, Germany) and lignin (alkaline, Sigma-Aldrich, Steinheim, Germany) in the proportion described in Supplemental Table S2 (Adapa et al. 2011). The model displayed R2 values of 0.9876, 0.9889, and 0.9763 and a slope of 0.9788, 1.000, and 0.9987 for hemicellulose, cellulose, and lignin, respectively. These models were then used to infer the proportion of each component in the samples (FitzPatrick et al. 2012; Krasznai et al. 2012). Finally, the degradation of hemicellulose, cellulose, and lignin was estimated by subtracting the percentage of the residual substrate from the total percentage of each hemicellulose component before degradation. Degradation rate was calculated using the following equation: \( \frac{Ci- Cf}{Ci}x100 \), where Ci is the total amount of compound before degradation and Cf is the residual component after degradation (Wang et al. 2011).
Statistical analyses
One-way analysis of variance (ANOVA) followed by Tukey HSD pairwise group comparisons was performed in IBM SPSS Statistics version 24 (SPSS Inc., Chicago, USA).
Results
Halotolerant lignocellulolytic consortia are capable of degrading lignocellulose biomass under high-salt conditions
The microbial community from the salt marsh soil, used as the inoculum, was able to adapt to, and grow on, wheat straw as the single carbon and energy source and under saline conditions. Using microscopic counts, we found that, during the adaptation phase, from transfer one to six, the cultures exhibited a progressively increasing fitness, as indicated by an increasing specific growth rate over time. The average specific growth rate µ (h−1; ± standard deviation; see Fig. 1a) increased from 0.22 h−1 (± 0.01) to 0.70 h−1 (± 0.03), from T1 to T6. In the stabilization phase, we observed an almost twofold reduction in the growth rate immediately after substrate change, which dropped from 0.70 h−1 (±0.03) to 0.38 h−1 (±0.02) (Fig. 1a, see T6 and T7), after which it remained constant until the end of the experiment (T10). The reduced apparent fitness of the consortia was thus related to the increased recalcitrance of the substrate.
Microbial growth rates and abundances during the enrichment. a Specific growth rate µ (day−1) of microbial communities across the enrichment processes, as determined by microscopic cell counts. b Bacterial abundances during the enrichment (log copies per mL), as determined by qPCR targeting the 16S rRNA gene. c Fungal abundances during the enrichment (log copies per mL), as determined by qPCR targeting the ITS1 region. Yellow bars—original soil inoculum; blue circles and bars—adaption phase using fresh lignocellulose substrate (transfer 1 to 6); red diamonds and bars—stabilization phase using pre-digested substrate (transfer 7 to 10). Bars refer to standard errors of the mean (n = 3)
The microscopic cell counts were corroborated by the 16S rRNA gene and ITS1 copy numbers determined by qPCR, which were used as proxies for bacterial (Fig. 1b) and fungal community density (Fig. 1c), respectively. At the end of each transfer in the adaptation phase, the consortia reached maximal bacterial levels of (log scale): 7.5 ± 1.3 (T1), 9.1 ± 0.002 (T3), and 9.2 ± 0.034 (T6) (average log 16S rRNA gene copies per mL ± standard deviation). In the stabilization phase, these values were similar: 9.2 ± 0.034 (T7) and 9.1± 0.02 (T10). The fungal abundances (measured by numbers of ITS1 gene copies) at the end of transfers 1, 3, 6, and 7 reached around (log scale) 6 per mL. However, we observed a significant reduction of ITS1 copies in the stabilization phase, from T7 to T10 (T test, P < 0.05), indicating that under saline conditions, fungi were strongly deselected by the increase of substrate recalcitrance.
Shifts in bacterial and fungal community composition
The microbial consortia were first analyzed by bacterial- as well as fungal-specific PCR-DGGE to examine the overall changes in community composition in selected transfers. Multidimensional scaling (MDS) of the bacterial community composition indicated a clear separation between the inoculum and the enriched communities and revealed the existence of two different clusters, separated on the basis of growth on fresh (adaptation phase) versus recalcitrant substrate (stabilization phase) (PERMANOVA, P < 0.05, Supplemental Fig. S3; Supplemental Fig. S4).
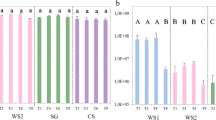
Lignocellulose degradation potential of the communities enriched during the experiment. Percentage reduction of hemicellulose, cellulose, and lignin contents of wheat straw (substrate) comparing with substrate recovered from an not uninoculated control. Explanation: 100% lignin, 100% cellulose, and 100% hemicellulose are equivalent at 18.3% of lignin, 42.5% of cellulose, and 32.5% of hemicellulose in the substrate respectively. Bars refer to standard errors of the mean (n = 3)
In contrast, the fungal consortia did not reveal a strong clustering between adaptation and stabilization phases, although they were significantly different from each other (PERMANOVA, P < 0.05) (Supplemental Fig. S4). The change in fungal community composition in the stabilization phase was associated with a substantial reduction of the number of bands, confirming the previously described qPCR results, which indicated that, under the applied conditions, fungi are deselected and outcompeted by bacteria.
Degradation of wheat straw by the microbial consortia
All consortia were found to preferably consume the hemicellulose part of the substrate, which was up to 80% degraded (Fig. 2). None of the selected consortia presented significant differences in hemicellulose degradation (ANOVA, P > 0.05). Interestingly, the cellulose part of the wheat straw was degraded to a lower extent, i.e., slightly above 40% (Fig. 2). Comparisons between the consortia across time indicated there was no significant difference in the degradation of hemicellulose, cellulose, and lignin (ANOVA, P > 0.05), except at T7 and T10, at which time points significant differences in the degradation of cellulose and lignin were found. The consortia at T10 degraded significantly more cellulose (64.2% ± 6.6) and lignin (61.4% ± 5.7) than those at T7 (cellulose 47% ± 10.8 and lignin 47.8% ± 6.6; ANOVA, P < 0.05) (Fig. 2). Comparing the two phases, the consortia from the stabilization phase were able to degrade significantly more lignin than those from the adaptation phase (T test, P < 0.05).
Communities structure of the degrading consortia, as determined by 16S rRNA gene-based sequencing
Direct amplicon sequencing performed on a selected number of transfers revealed grossly decreasing bacterial richness values along the transfers. Specifically, for the inocula and the T1, T3, T6, T7, and T10 consortia, the values were 4.84 ± 0.34, 3.49 ± 0.40, 3.40 ± 0.72, 3.14 ± 0.25, 3.41 ± 0.38, and 2.90 ± 0.27, respectively (log OTU number ± standard deviation). Moreover, significant differences in richness were found between the consortia in the adaptation and the stabilization phases, T1, T3 and T6 versus T7 and T10, respectively (ANOVA, P < 0.05).
Regarding the bacterial community structures (β-diversities), PCoA of the unweighted UniFrac community distances confirmed the previously described PCR-DGGE results. The data showed that the consortia selected on fresh substrate (adaptation phase, T1, T3, and T6) were markedly different from those selected on recalcitrant substrate (T7 and T10) (Fig. 3). PERMANOVA showed that, indeed, bacterial consortia were significantly different between the adaptation and stabilization phases, as driven by the change in the substrate (P < 0.005). This indicated that a clear shift had occurred as a result of the transition from raw to recalcitrant substrate.
Shifts in bacterial community structure during the adaptation and stabilization phases of the experiment as derived from the 16S rRNA gene sequencing data (V4-V5 region). Principal coordinates analysis (PCoA) of unweighted UniFrac distances for 16S rRNA gene sequencing data of selected enrichment consortia (T1, T3, T6, T7, T10). Fresh substrate (blue circles), used substrate (red diamonds), inoculum (green asterisks). PERMANOVA indicated significant differences between the communities (P = 0.007, Pseudo-F = 2.90)
The comparison of the bacterial consortia between the transfers showed that, in the adaptation phase, a large amount of OTUs was significantly affected by the enrichment, leading to a large turnover in community composition and positive selection of OTUs. In contrast, the turnover was lower in the stabilization phase, with relatively few OTUs being negatively affected by the confrontation with the recalcitrant substrate (T7) (Fig. 4). Comparison of the consortia at T7 and T10 (stabilization phase) revealed an increase of abundance of particular OTUs (Fig. 4). Thus, 19 OTUs were differentially selected in the adaptation phase (Table 1) and only five OTUs were positively affected by the change in the substrate during stabilization phase (Table 2). Four OTUs were present in both phases: OTU57506 (affiliated with Halomonas alkaliphila), OTU415 (affiliated with Algoriphagus winogradskyi or ratkowskyi, OTU358 (Joostella marina), and OTU667 (Flavobacterium beibuense).
Number of OTUs (log2 fold change) that were positively and negatively influenced in the adaptation and stabilization phases of the experiment. DESeq2 function for phyloseq was used to obtain the statistically significant OTUs affected by the enrichment process and the change in substrate composition. Comparisons between selected transfers for adaptation phase included inoculum vs T1, T1 vs T3, and T3 vs T6 (blue squares). In the stabilization phase, the comparison was made between T3 vs T6 and T7 vs T10 (red squares). The adaptation phase shows an important reduction of numbers of OTUs, as indicated by a larger number of bars with negative values especially in the early and late transfers, whereas in the stabilization phase, we observed an increase in the number of OTUs selected—mostly OTUs with positive values were significantly different from one transfer to another
Degradation of wheat straw by selected strains
In total, 47 bacterial strains were recovered from the consortia at T6 and T10. Most of the strains were isolated from both transfers, except for Photobacterium halotolerans A34, Albirhodobacter marinus C13, and Paracoccus seriniphilus C14, which were recovered only from the adaptation phase (T6) (Table 3). All were identified on the basis of 16S rRNA gene sequencing (Tables 3 and 4). Subsequently, bacterial strains were screened for the production of enzymes able to degrade X-glu, X-cell, X-gal, X-xyl, X-man and X-fuc (Tables 3 and 4). The data showed that such degradation potential was widespread across the strains. Of the 47 strains tested, only three did not show any enzymatic activity against the selected substrates. These were Staphylococcus capitis P1, Bacillus oleronius G13, and Erythrobacter gaetbuli G57.
By aligning the 16S rRNA gene sequences recovered from the isolated bacteria with those of the OTUs obtained by direct sequencing (Fig. 5), we were able to pinpoint the strains that were highly abundant in the consortia (Tables 3 and 4). In the adaptation phase, nine strains were closely related to four enriched OTUs (Table 3). Those were affiliated with Halomonas alkaliphila (M10 and M11), Photobacterium halotolerans (A34, M14, M15, and M20), Paracoccus seriniphilus (C14 and M48), and Altererythrobacter indicus (P4, G10, and G19). In the stabilization phase, seven strains were closely related to four enriched OTUs (Table 4): Halomonas meridiana M11, Algoriphagus winogradskyi G63, Jootella marina (G54, G65, and ME32), and Flavobacterium beibuense (M35 and M44). Finally, we recovered two strains affiliated with Pseudomonas sabulinigri G20 and M7; however, these did not match the OTU665 (affiliated with Pseudomonas putida) (Fig. 5).
Phylogenetic affiliation of 16S rRNA gene sequences of isolated strains and sequenced OTUs. Neighbor Joining tree based on the 16S rRNA gene sequences (V4-V5 region) from bacterial strains and from the significant abundant OTUs at the end of the adaptation phase (T6) and stabilization phase (T10). For the adaptation phase, underlined in blue are OTU850 H. alkaliphila (99%, MF928383.1), OTU859 P. halotolerans (99%, KT354559.1), OTU176 P. seriniphilus (99%, KX453219.1), and OTU659, Altererythrobacter sp. (99%, KT325206.1). For the stabilization phase, underlined in red, OTU57506 H. meridiana (99%), OTU665 P. putida (99%), OTU415 A. winogradskyi/ratkowskyi (98%), OTU358 J. marina (99%, DQ768627.1), and OTU667 F. beibuense (99%, KY819115.1). Between brackets: % of identity, reference accession number. The 16S rRNA gene sequence from Methanocaldococcus jannaschii was used as outgroup. Bar indicated divergence scale (0.2 = 20%)
Tables 3 and 4 show details of enzyme production by the strains. On the one hand, strains isolated from the adaptation phase yielded not only most of the tested hydrolytic activities, but also showed the highest activities. Remarkably, the strains affiliated with Microbacterium oleivorans (G37, G46) and Devosia psychrophila (G33-G35) revealed the production of five or even six hydrolytic enzymes (Table 3). On the other hand, strains isolated from the recalcitrant substrate were less versatile than those isolated from fresh substrate, as evidenced by the lower number of enzymatic activities (three out of six tested). Only the strains affiliated with J. marina (ME32, G54, and G65) presented the capacity to produce at least four hydrolytic enzymes with high activity.
Fungal strains from the stabilization phase
As mentioned before, the change in the substrate had an important effect on the fungal community. Only one fungal strain was obtained from the adaptation phase (Table 3). It was affiliated with Sarocladium strictum HF1 and was obtained from all triplicate plates. It was, however, not possible to recover any fungal strain from the stabilization phase. Despite the bands observed in DGGE (based on the ITS1 region) in the stabilization phase, we observed a sharp decline in fungal abundance—as determined by qPCR targeting the same region (Fig. 1b)—at the end of the experiment (T10), which probably hindered isolation.
Discussion
In this study, we produced and characterized microbial consortia—potential sources of lignocellulose degraders and their enzymes—that were capable of degrading wheat straw under high-salt concentrations, a condition often established by particular lignocellulose pretreatment steps. Thus, our selected halotolerant microbial consortia represent a clear prospect of lignocellulose degradation under saline conditions, as they may either be used to directly unlock lignocellulose biomass or to produce halotolerant lignocellulolytic enzymes. The latter application may eliminate the expensive washing steps, reducing costs.
Saline conditions favor bacterial over fungal degraders
Changes in wheat straw content can considerably affect the composition of microbial communities growing on it. Here, in particular, fungal densities decreased significantly in the stabilization phase (when recalcitrant substrate was used), hindering our ability to isolate fungal strains. It is generally believed that fungi are ubiquitous and capable of occupying virtually every ecological niche as a result of their ability to degrade a suite of organic compounds such as complex biological polymers. They may also play roles in degrading lignocellulose in marine environments (Richards et al. 2012), where the major factors affecting their diversities are salt concentration and temperature (Fuentes et al. 2015). For instance, it has been shown that several fungal strains recovered from mangrove systems are capable of growing on wood under high-salt conditions (Arfi et al. 2013). In our study, the only isolated fungal strain—Sarocladium strictum, previously known as Acremonium strictum (Summerbell et al. 2011)—is likely well adapted to saline environments, as it was previously isolated from a marine ecosystem (Fuentes et al. 2015). Here it originated from a salt-marsh soil inoculum. It was, however, only recovered in the adaptation phase, declining in density (to below the detection limit) in the stabilization part of the experiment. Although we cannot pinpoint the exact reason for the observed decline in fungal density (considering that both temperature and salt concentration remained constant in our experiment), we argue that this reduction could be explained by nitrogen depletion in the recalcitrant substrate, consistent with the findings by Meidute et al. (2008). Thus, the impossibility to isolate fungal strains from the specialized consortia could be related to a very strong nutritional demand under the prevailing conditions, leading to a decline in density that hindered isolation. Additionally, pH could be an important factor affecting the viability of the fungi in our system. During the cultivation, the pH decreased slightly from 7.2 to 6.8, which is higher than the optimal pH for fungal growth (between pH 2.2 and 6.5; Matthies et al. 1997). Moreover, the maintenance of the almost neutral pH along the incubation suggested a low production of organic acids (which indicates that massive fermentation did not occur). The maintenance of prevailing aerobic conditions in the culture probably incited mostly oxidative phosphorylation processes. Thus, in the system (an agitated saline environment with a recalcitrant source of carbon and energy), bacteria probably had a main role in the degradation process.
The dominance of bacteria over fungi in our halotolerant lignocellulose grown consortia is interesting, as previous studies, performed under non-saline conditions, suggested that fungal communities have a relevant participation in lignocellulose degradation, even working in liquid and agitated systems. For example, Brossi et al. (2015) found that C. ligniaria (strains WS1, WS2, SG8) had a significant role in the degradation of diverse lignocellulose feedstocks, while Jiménez et al. (2013) found the same organism (strain 2w1F) played a crucial role in the decomposition of wheat straw in presence of 5-hydroxymethylfurfural. In both cases, the dilution-to-stimulation approach was used for the selection of the degrader communities.
Substrate quality greatly impacts community composition
The findings in this study clearly indicate that substrate quality and composition direct the structure of microbial consortia (Simmons et al. 2014; Brossi et al. 2015), which developed to degrade either fresh (adaptation phase) or previously- degraded (recalcitrant) lignocellulose substrate (stabilization phase). Whereas the fresh substrate allowed the selection of a more generalist degrading community, composed of very specific bacterial and fungal strains, the recalcitrant substrate selected for more specialized, mostly bacterial, species. Interestingly, all replicates of the enrichment process gave fairly similar patterns, in terms of consortium development, both quantitatively (viz the bacterial and fungal abundance values) and with respect to the bacterial community structures, demonstrating the robustness of our findings. We thus posit here that a consistent selection of microorganisms with progressively higher abilities to grow (jointly) on the substrate had taken place. In the consortia, bacteria were quantitatively by far more important than fungi, and so we placed a greater focus on the bacterial part of the resulting consortia. This bacterial dominance was even exacerbated by the shift to a more recalcitrant substrate after T6.
Wheat straw degradation and potential involvement of identified strains
On the basis of all our data, we depict the degradation of wheat straw under saline conditions to proceed in a sequential manner, with different microbes being dominant in a spatiotemporally explicit form. The wheat straw, being recalcitrant, poses clear obstacles to degradation. The main hurdles are the presence of crystalline cellulose and the bonding between lignin and hemicellulose (shielding the latter component from access by key enzymes). We briefly discuss these issues in the paragraphs below.
Crystalline cellulose is highly recalcitrant to chemical and biological hydrolysis due to the strongly linked chains of cellodextrins. The decomposition of crystalline cellulose, for example filter paper, requires the production of specific cellulases. In our consortia, Joostella marina (OTU358) and Flavobacterium beibuense(OTU667) may have had a main role in cellulose degradation, as we observed increases in their abundances in the stabilization phase. Also, the consortia from this phase displayed higher cellulose degradation capacities than consortia from the adaptation phase. Both J. marina (OTU358) and F. beibuense (OTU667) belong to the Flavobacteriaceae (Bernardet et al. 2002). Members of this family have been isolated from soil, sediment and marine/saline environments, and they have been typically associated with decomposition of complex polysaccharides (Lambiase 2014). Some species in the family degrade soluble cellulose derivatives such as carboxymethyl-cellulose. However, since enzymes other than cellulases can degrade this compound, this does not demonstrate that these species are cellulolytic. J. marina probably has an important role in the degradation of recalcitrant regions of lignocellulose substrate, as it is capable to grow on complex hydrocarburic substrate (Rizzo et al. 2015). The organism is strictly aerobic and can grow in up to 15% NaCl, with glucose, arabinose, mannose, and cellobiose as single carbon and energy sources. Additionally, it has been reported to be positive for α-glucosidase, β-glucosidase, β-galactosidase, and α-mannosidase production (Stackebrandt et al. 2013). In our final consortia, J. marina could be associated with the degradation of the crystalline cellulose in the wheat straw. However, more studies are needed to demonstrate such cellulolytic capability. Currently, this characteristic is restricted to members of the Cytophagaceae family (Bernardet et al. 2002). Additionally, the Flavobacterium species found in this study (F. beibuense OTU667 and Flavobacterium suzhouense OTU496) may be only associated with the degradation of amorphous cellulose, which is readily digestible. These organisms can degrade soluble cellulose such as hydroxymethylcellulose and cellodextrine (Lambiase 2014).
Regarding lignin degradation or bond hydrolysis, the increasing abundance of Pseudomonas species (P. putida OTU665 and P. sabulinigri G20, M38, and M7) in the stabilization phase suggest a role for these organisms in the relevant transformation steps, such as the degradation of recalcitrant regions of the substrate like residual hemicellulose linked to lignin structures. Pseudomonas species stand out as having a great potential capacity for lignin degradation (Beckham et al. 2016). For instance, in a recent study, P. monteilli and P. plecoglossicida were enriched from mature vegetal compost. These organisms were found to degrade a large amount of lignin-related compounds (Ravi et al. 2017). In another study, Salvachúa et al. (2015) isolated P. putida, Rhodococcus jostii, and Acinetobacter sp. ADP1, all of which were able to depolymerize and catabolize high-molecular-weight lignin (Salvachúa et al. 2015).
The most labile part of the substrate, hemicellulose, was probably mainly attacked by H. meridiana (OTU 57506) and related species. Their decreased abundance in the stabilization phase could indicate that the hemicellulose part of the substrate was largely depleted. H. meridiana belongs to the class Gammaproteobacteria. It is a facultatively halotolerant organism capable of growth in NaCl concentrations between 0.1 and 32.5% (w/v). It is mostly found in marine environments (Octavia and Lan 2014). A recent study suggested that H. meridiana has great potential for biotechnology applications, as a producer of extracellular enzymes adapted to salinity (Yin et al. 2015).
Finally, Algoriphagus winogradskyi/ratkowskyi G63, belonging to the Cytophagaceae, could be involved in the degradation of both the hemicellulose and cellulose regions of the substrate. A genetic analysis of Algoriphagus sp. PR1 demonstrated its high capacity of polysaccharide degradation, as large numbers of genes encoding glycoside hydrolases, polysaccharide lyases, carbohydrate esterases, and glycosyltransferases were found (Alegado et al. 2011). Previous reports indicated that related strains cannot degrade filter paper (Lambiase 2014), however our strains were not yet tested for such activity.
Although the contribution of fungi to the degradation process seems to be restricted to the adaptation phase, previous reports have demonstrated the biotechnological application of Sarocladium strictum. Interestingly, this was our only isolated fungal strain, and one may envision a role for it in the production of cellulases direct from infested lignocellulose feedstock (Goldbeck et al. 2013). Also, a gene for gluco-oligosaccharide oxidase from this species has been engineered (high catalytic activity and lowsubstrate inhibition) for application in industrial plant polysaccharide degradation (Domon et al. 2013). Definitely, more studies are necessary on S. strictum to examine all its degradation capacities, although it might be restricted to conditions with high nutrient supply.
In conclusion, the construction of microbial consortia able to grow on wheat straw as a carbon and energy source under saline conditions offers access to salt-adapted or salt-tolerant enzymes (haloenzymes) that enable the development of processes under saline conditions. It is assumed that the selected organisms harbor the potential to naturally produce such salt-adapted enzymes, which are applicable in a bioprocess with raised NaCl levels. We propose that the key members of our consortia yield very interesting salt-tolerant enzymes for bioengineering, as follows: (1) J. marina (G54, G65, ME32): production of carbohydrate esterases, (2) F. beibuense (M35, M44): production of cellulases, (3) P. sabulinigri (G20, M38, M7): production of ligninases, and (4) H. meridiana (G21): production of hemicellulases. A key issue here is the precise combination of enzymes that is required to establish an efficient “saline bioprocess”. Potentially, such an enzyme mixture is made on the basis of the organisms as described here.
References
Adapa PK, Schonenau LG, Canam T, Dumonceaux T (2011) Quantitative analysis of lignocellulosic components of non-treated and steam exploded barley, canola, oat and wheat straw using Fourier transform infrared spectroscopy. J Agric Sci Technol 1:177–188
Alegado RA, Ferriera S, Nusbaum C, Young SK, Zeng Q, Imamovic A, Fairclough SR, King N (2011) Complete genome sequence of Algoriphagus sp. PR1, bacterial prey of a colony-forming Choanoflagellate. J Bacteriol 193(6):1485–1486. https://doi.org/10.1128/JB.01421-10
Arfi Y, Chevret D, Henrissat B, Berrin J-G, Levasseur A, Record E (2013) Characterization of salt-adapted secreted lignocellulolytic enzymes from the mangrove fungus Pestalotiopsis sp. Nat Commun 4:1810. https://doi.org/10.1038/ncomms2850
Beckham GT, Johnson CW, Karp EM, Salvachúa D, Vardon DR (2016) Opportunities and challenges in biological lignin valorization. Curr Opin Biotechnol 42:40–53. https://doi.org/10.1016/j.copbio.2016.02.030
Bernardet J-FO, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52(3):1049–1070. https://doi.org/10.1099/ijs.0.02136-0
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10(1):57–59. https://doi.org/10.1038/nmeth.2276
Brethauer S, Studer MH (2015) Biochemical conversion processes of lignocellulosic biomass to fuels and chemicals—a review. Chim Int J Chem 69(10):572–581. https://doi.org/10.2533/chimia.2015.572
Brons JK, van Elsas JD, (2008) Analysis of bacterial communities in soil by use of denaturing gradient gel electrophoresis and clone libraries, as influenced by different reverse primers. App Environ Microbiol 74:2717–2727
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6(8):1621–1624. https://doi.org/10.1038/ismej.2012.8
Cortes-Tolalpa L, Jiménez DJ, Brossi MJ de L, Salles JF, van Elsas JD (2016) Different inocula produce distinctive microbial consortia with similar lignocellulose degradation capacity. Appl Microbiol Biotechnol 100(17):7713–7725. https://doi.org/10.1007/s00253-016-7516-6
Brossi MJ de L, Jiménez DJ, Cortes-Tolalpa L, van Elsas JD (2015) Soil-derived microbial consortia enriched with different plant biomass reveal distinct players acting in lignocellulose degradation. Microb Ecol 71(3):616–627. https://doi.org/10.1007/s00248-015-0683-7
Dini-Andreote F, Pereira e Silva MC, Triado-Margarit X, Casamayor EO, van Elsas JD, Salles JF (2014) Dynamics of bacterial community succession in a salt marsh chronosequence: evidences for temporal niche partitioning. ISME J 8(10):1989–2001. https://doi.org/10.1038/ismej.2014.54
Domon B, Costello CE, Foumani M, Juvonen M, Seppälä J, Tenkanen M, Master ER, Tuomainen P, Saulnier L, Tenkanen M (2013) A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J 5(4):397–409. https://doi.org/10.1007/BF01049915
FitzPatrick M, Champagne P, Cunningham MF (2012) Quantitative determination of cellulose dissolved in 1-ethyl-3-methylimidazolium acetate using partial least squares regression on FTIR spectra. Carbohydr Polym 87(2):1124–1130. https://doi.org/10.1016/j.carbpol.2011.08.086
Fuentes ME, Quiñones RA, Gutiérrez MH, Pantoja S (2015) Effects of temperature and glucose concentration on the growth and respiration of fungal species isolated from a highly productive coastal upwelling ecosystem. Fungal Ecol 13:135–149. https://doi.org/10.1016/j.funeco.2014.09.006
Goldbeck R, Ramos MM, Pereira GAG, Maugeri-Filho F (2013) Cellulase production from a new strain Acremonium strictum isolated from the Brazilian biome using different substrates. Bioresour Technol 128:797–803. https://doi.org/10.1016/j.biortech.2012.10.034
Gunny AAN, Arbain D, Edwin Gumba R, Jong BC, Jamal P (2014) Potential halophilic cellulases for in situ enzymatic saccharification of ionic liquids pretreated lignocelluloses. Bioresour Technol 155:177–181. https://doi.org/10.1016/j.biortech.2013.12.101
Jiménez DJ, Korenblum E, van Elsas JD (2013) Novel multispecies microbial consortia involved in lignocellulose and 5-hydroxymethylfurfural bioconversion. Appl Microbiol Biotechnol 98(6):2789–2803. https://doi.org/10.1007/s00253-013-5253-7
Jönsson LJ, Martin C (2016) Pretreatment of lignocellulose: formation of inhibitory by-products and strategies for minimizing their effects. Bioresour Technol 199:103–112. https://doi.org/10.1016/j.biortech.2015.10.009
Khoo HH, Ee WL, Isoni V (2016) Bio-chemicals from lignocellulose feedstock: sustainability, LCA and the green conundrum. Green Chem 18(7):1912–1922. https://doi.org/10.1039/C5GC02065D
Kinet R, Destain J, Hiligsmann S, Thonart P, Delhalle L, Taminiau B, Daube G, Delvigne F (2015) Thermophilic and cellulolytic consortium isolated from composting plants improves anaerobic digestion of cellulosic biomass: toward a microbial resource management approach. Bioresour Technol 189:138–144. https://doi.org/10.1016/j.biortech.2015.04.010
Krasznai DJ, Champagne P, Cunningham MF (2012) Quantitative characterization of lignocellulosic biomass using surrogate mixtures and multivariate techniques. Bioresour Technol 110:652–661. https://doi.org/10.1016/j.biortech.2012.01.089
Lambiase A (2014) The family Sphingobacteriaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The prokaryotes other major lineages of bacteria and the archaea, 4th edn. Springer Berlin Heidelberg, Berlin, pp 907–914
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550–571. https://doi.org/10.1186/s13059-014-0550-8
Maruthamuthu M, Jiménez DJ, Stevens P, van Elsas JD (2016) A multi-substrate approach for functional metagenomics-based screening for (hemi)cellulases in two wheat straw-degrading microbial consortia unveils novel thermoalkaliphilic enzymes. BMC Genomics 17(1):86. https://doi.org/10.1186/s12864-016-2404-0
Matthies C, Erhard H-P, Drake HL (1997) Effects of pH on the comparative culturability of fungi and bacteria from acidic and less acidic forest soils. J Basic Microbiol 37(5):335–343. https://doi.org/10.1002/jobm.3620370506
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8(4):e61217. https://doi.org/10.1371/journal.pone.0061217
Mcmurdie PJ, Holmes S, Mchardy AC (2014) Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531
Meidute S, Demoling F, Bååth E (2008) Antagonistic and synergistic effects of fungal and bacterial growth in soil after adding different carbon and nitrogen sources. Soil Biol Biochem 40(9):2334–2343. https://doi.org/10.1016/j.soilbio.2008.05.011
Octavia S, Lan R (2014) The family Enterobacteriaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson FL (eds) The prokaryotes: Gammaproteobacteria, 4th edn. Springer Berlin Heidelberg, Berlin, pp 225–286
Okeke BC, Lu J (2011) Characterization of a defined cellulolytic and xylanolytic bacterial consortium for bioprocessing of cellulose and hemicelluloses. Appl Biochem Biotechnol 163(7):869–881. https://doi.org/10.1007/s12010-010-9091-0
Pereira e Silva MC, Dias ACF, van Elsas JD, Salles JF (2012) Spatial and temporal variation of archaeal, bacterial and fungal communities in agricultural soils. PLoS One 7(12):e51554. https://doi.org/10.1371/journal.pone.0051554
Rabemanolontsoa H, Saka S (2016) Various pretreatments of lignocellulosics. Bioresour Technol 199:83–91. https://doi.org/10.1016/j.biortech.2015.08.029
Ravi K, Garcia-Hidalgo J, Gorwa-Grauslund MF, Lidén G (2017) Conversion of lignin model compounds by Pseudomonas putida KT2440 and isolates from compost. Appl Microbiol Biotechnol 101(12):5059–5070. https://doi.org/10.1007/s00253-017-8211-y
Richards TA, Jones MDM, Leonard G, Bass D (2012) Marine fungi: their ecology and molecular diversity. Annu Rev Mar Sci 4(1):495–522. https://doi.org/10.1146/annurev-marine-120710-100802
Rideout JR, He Y, Navas-Molina JA, Walters WA, Ursell LK, Gibbons SM, Chase J, McDonald D, Gonzalez A, Robbins-Pianka A, Clemente JC, Gilbert JA, Huse SM, Zhou H-W, Knight R, Caporaso JG (2014) Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. Peer J 2:e545. https://doi.org/10.7717/peerj.545
Rizzo C, Michaud L, Graziano M, De Domenico E, Syldatk C, Hausmann R, Lo Giudice A (2015) Biosurfactant activity, heavy metal tolerance and characterization of Joostella strain A8 from the Mediterranean polychaete Megalomma claparedei (Gravier, 1906). Ecotoxicology 24(6):1294–1304. https://doi.org/10.1007/s10646-015-1504-y
Salvachúa D, Karp EM, Nimlos CT, Vardon DR, Beckham GT (2015) Towards lignin consolidated bioprocessing: simultaneous lignin depolymerization and product generation by bacteria. Green Chem 17(11):4951–4967. https://doi.org/10.1039/C5GC01165E
Simmons CW, Reddy AP, Simmons BA, Singer SW, VanderGheynst JS (2014) Effect of inoculum source on the enrichment of microbial communities on two lignocellulosic bioenergy crops under thermophilic and high-solids conditions. J Appl Microbiol 117(4):1025–1034. https://doi.org/10.1111/jam.12609
Stackebrandt E, Chertkov O, Lapidus A, Nolan M, Lucas S, Han C, Cheng J-F, Tapia R, Goodwin LA, Bruce D, Pitluck S, Liolios K, Mavromatis K, Pagani I, Ivanova N, Mikhailova N, Huntemann M, Pati A, Chen A, Palaniappan K, Rohde M, Tindall BJ, Göker M, Woyke T, Detter JC, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Klenk H-P, Kyrpides NC (2013) High-quality-draft genome sequence of the yellow-pigmented flavobacterium Joostella marina type strain (En5(T)). Stand Genomic Sci 8(1):37–46. https://doi.org/10.4056/sigs.3537045
Summerbell RC, Gueidan C, Schroers H-J, de Hoog GS, Starink M, Rosete YA, Guarro J, Scott JA (2011) Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud Mycol 68:139–162. https://doi.org/10.3114/sim.2011.68.06
Sun S, Sun S, Cao X, Sun R (2016) The role of pretreatment in improving the enzymatic hydrolysis of lignocellulosic materials. Bioresour Technol 199:49–58. https://doi.org/10.1016/j.biortech.2015.08.061
Talebnia F, Karakashev D, Angelidaki I (2010) Production of bioethanol from wheat straw: an overview on pretreatment, hydrolysis and fermentation. Bioresour Technol 101(13):4744–4753. https://doi.org/10.1016/j.biortech.2009.11.080
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Wang W, Yan L, Cui Z, Gao Y, Wang Y, Jing R (2011) Characterization of a microbial consortium capable of degrading lignocellulose. Bioresour Technol 102(19):9321–9324. https://doi.org/10.1016/j.biortech.2011.07.065
Wang M, Yang P, Falcão Salles J (2016) Distribution of root-associated bacterial communities along a salt-marsh primary succession. Front Plant Sci 6:1188. https://doi.org/10.3389/fpls.2015.01188
Xu F, Yu J, Tesso T, Dowell F, Wang D (2013) Qualitative and quantitative analysis of lignocellulosic biomass using infrared techniques: a mini-review. Publ from USDA-ARS / UNL Fac 104:801–809
Yin J, Chen J-C, Wu Q, Chen G-Q (2015) Halophiles, coming stars for industrial biotechnology. Biotechnol Adv 33:1433–1442. https://doi.org/10.1016/j.biotechadv.2014.10.008
Acknowledgements
This work was supported by Consejo Nacional de Ciencia y Tecnología (CONACyT) through the PhD scholarship to Larisa Cortes Tolalpa and NWO grant (Microwaste project) to Joana Falcao Salles. We would like to thank Jolanda Brons for her technical support, as well as Paul Dockerty, Fernando Guzman Chavez, Leonel Herrera Alsina, and Maryam Chaib the Mares for their support and valuable comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human studies and informed consent
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 587 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Cortes-Tolalpa, L., Norder, J., van Elsas, J.D. et al. Halotolerant microbial consortia able to degrade highly recalcitrant plant biomass substrate. Appl Microbiol Biotechnol 102, 2913–2927 (2018). https://doi.org/10.1007/s00253-017-8714-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8714-6