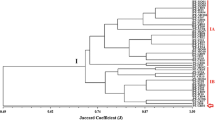
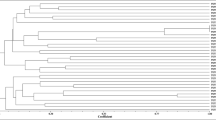
Abstract
In the present study, the relative distribution of endophytic rhizobia in field-collected root nodules of the promiscuous host mung bean was investigated by sequencing of 16S ribosomal RNA (rRNA) and nifH genes, amplified directly from the nodule DNA. Co-dominance of the genera Bradyrhizobium and Ensifer was indicated by 32.05 and 35.84% of the total retrieved 16S rRNA sequences, respectively, and the sequences of genera Mesorhizobium and Rhizobium comprised only 0.06 and 2.06% of the recovered sequences, respectively. Sequences amplified from rhizosphere soil DNA indicated that only a minor fraction originated from Bradyrhizobium and Ensifer strains, comprising about 0.46 and 0.67% of the total retrieved sequences, respectively. 16S rRNA gene sequencing has also identified the presence of several non-rhizobial endophytes from phyla Proteobacteria, Actinobacteria, Bacteroides, and Firmicutes. The nifH sequences obtained from nodules also confirmed the co-dominance of Bradyrhizobium (39.21%) and Ensifer (59.23%) strains. The nifH sequences of the genus Rhizobium were absent, and those of genus Mesorhizobium comprised only a minor fraction of the sequences recovered from the nodules and rhizosphere soil samples. Two bacterial isolates, identified by 16S rRNA gene sequence analysis as Bradyrhizobium strain Vr51 and Ensifer strain Vr38, successfully nodulated the original host (mung bean) plants. Co-dominance of Bradyrhizobium and Ensifer strains in the nodules of mung bean indicates the potential role of the host plant in selecting specific endophytic rhizobial populations. Furthermore, successful nodulation of mung bean by the isolates showed that strains of both the genera Bradyrhizobium and Ensifer can be used for production of inoculum.







Similar content being viewed by others
References
Aserse AA, Räsänen LA, Aseffa F, Hailemariam A, Lindström K (2013) Diversity of sporadic symbionts and nonsymbiotic endophytic bacteria isolated from nodules of woody, shrub, and food legumes in Ethiopia. Appl Microbiol Biotechnol 97:10,117–10,134
Ayyaz K, Zaheer A, Rasul G, Mirza MS (2016) Isolation and identification by 16S rRNA sequence analysis of plant growth-promoting azospirilla from the rhizosphere of wheat. Braz J Microbiol 47:542–550. https://doi.org/10.1016/j.bjm.2015.11.035
Bhattacharyya P, Jha D (2012) Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J Microbiol Biotechnol 28:1327–1350
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen A, McGarrell DM, Marsh T, Garrity GM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
De Meyer SE, De Beuf K, Vekeman B, Willems A (2015) A large diversity of non-rhizobial endophytes found in legume root nodules in Flanders (Belgium). Soil Biol Biochem 83:1–11
Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853
Estrella MJ, Munoz S, Soto MJ, Ruiz O, Sanjuán J (2009) Genetic diversity and host range of rhizobia nodulating Lotus tenuis in typical soils of the Salado River basin (Argentina). Appl Environ Microbiol 75:1088–1098
Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37
Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, Cole JR (2013) FunGene: the functional gene pipeline and repository. Front Microbiol 4:1–14
Gtari M, Ghodhbane-Gtari F, Nouioui I, Beauchemin N, Tisa LS (2012) Phylogenetic perspectives of nitrogen-fixing actinobacteria. Arch Microbiol 194:3–11
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Sys Evol Microbiol 62:716–721
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1–5. https://doi.org/10.1093/molbev/msw054
Leite J, Fischer D, Rouws LF, Fernandes-Júnior PI, Hofmann A, Kublik S, Schloter M, Xavier GR, Radl V (2016) Cowpea nodules harbor non-rhizobial bacterial communities that are shaped by soil type rather than plant genotype. Front Plant Sci 7:2064
Li QQ, Wang ET, Zhang YZ, Zhang YM, Tian CF, Sui XH, Chen WF, Chen WX (2011) Diversity and biogeography of rhizobia isolated from root nodules of Glycine max grown in Hebei Province, China. Microb Ecol 61:917–931
Lu YL, Chen WF, Wang ET, Han LL, Zhang XX, Chen WX, Han SZ (2009) Mesorhizobium shangrilense sp. nov., isolated from root nodules of Caragana species. Int J Sys Evol Microbiol 59:3012–3018
Ma Y, Prasad M, Rajkumar M, Freitas H (2011) Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol Adv 29:248–258
Mirza BS, Muruganandam S, Meng X, Sorensen DL, Dupont RR, McLean JE (2014) Arsenic (V) reduction in relation to iron (III) transformation and molecular characterization of the structural and functional microbial community in sediments of a basin-fill aquifer in northern Utah. Appl Environ Microbiol 80:3198–3208
Mohammad W, Shehzadi S, Shah SM, Shah Z (2010) Effect of tillage and crop residues management on mungbean (Vigna radiata (L.) Wilczek) crop yield, nitrogen fixation and water use efficiency in rain fed areas. Pak J Bot 42:1781–1789
Oresnik IJ, Liu SL, Yost CK, Hynes MF (2000) Megaplasmid pRme2011a of Sinorhizobium meliloti is not required for viability. J Bacteriol 182:3582–3586
Pandya M, Kumar GN, Rajkumar S (2013) Invasion of rhizobial infection thread by non-rhizobia for colonization of Vigna radiata root nodules. FEMS Microbiol Lett 348:58–65
Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ (2015) Bacterial associations with legumes. Crit Rev Plant Sci 34:17–42
Pinto AJ, Raskin L (2012) PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS One 7:e43093
Poly F, Monrozier LJ, Bally R (2001) Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152:95–103
Risal CP, Djedidi S, Dhakal D, Ohkama-Ohtsu N, Sekimoto H, Yokoyama T (2012) Phylogenetic diversity and symbiotic functioning in mungbean (Vigna radiata L. Wilczek) bradyrhizobia from contrast agro-ecological regions of Nepal. Syst Appl Microbiol 35:45–53
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Sharma SB, Sayyed RZ, Trivedi MH, Gobi TA (2013) Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2:1–14
Simon Z, Mtei K, Gessesse A, Ndakidemi PA (2014) Isolation and characterization of nitrogen fixing rhizobia from cultivated and uncultivated soils of northern Tanzania. Am J Plant Sci 5:4050–4067
Singh G, Kumar D, Sharma P (2015) Effect of organics, biofertilizers and crop residue application on soil microbial activity in rice—wheat and rice-wheat mungbean cropping systems in the Indo-Gangetic Plains. Cogent Geosci 1:1085296
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wang Q, Quensen JF, Fish JA, Lee TK, Sun Y, Tiedje JM, Cole JR (2013) Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. MBio 4:e00592–e00513
Wilson K (1987) Preparation of genomic DNA from bacteria. In: Ausubel FM (ed) Current protocols in molecular biology. John Wiley&Sons, New York, United States, unit 2.4
Wright ES, Yilmaz LS, Noguera DR (2012) DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78:717–725
Yan J, Han XZ, Ji ZJ, Li Y, Wang ET, Xie ZH, Chen WF (2014) Abundance and diversity of soybean-nodulating rhizobia in black soil are impacted by land use and crop management. Appl Environ Microbiol 80:5394–5402
Yang JK, Yuan TY, Zhang WT, Zhou JC, Li YG (2008) Polyphasic characterization of mung bean (Vigna radiata L.) rhizobia from different geographical regions of China. Soil Biol Biochem 40:1681–1688
Zaheer A, Mirza BS, McLean JE, Yasmin S, Shah TM, Malik KA, Mirza MS (2016) Association of plant growth-promoting Serratia spp. with the root nodules of chickpea. Res Microbiol 167:510–520. https://doi.org/10.1016/j.resmic.2016.04.001
Zahran HH (1999) Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Biol Rev 63:968–989
Zahran HH (2017) Plasmids impact on rhizobia-legumes symbiosis in diverse environments. Symbiosis:1–17. https://doi.org/10.1007/s13199-017-0476-5
Zhang YF, Wang ET, Tian CF, Wang FQ, Han LL, Chen WF, Chen WX (2008) Bradyrhizobium elkanii, Bradyrhizobium yuanmingense and Bradyrhizobium japonicum are the main rhizobia associated with Vigna unguiculata and Vigna radiata in the subtropical region of China. FEMS Microbiol Lett 285:146–154
Zhang X-X, Tang X, Sheirdil RA, Sun L, Ma X-T (2014) Rhizobium rhizoryzae sp. nov., isolated from rice roots. Int J Sys Evol Microbiol 64:1373–1377
Acknowledgements
The provision of consumables by Pakistan Science Foundation through project PSF/NSLP/P-NIBGE (315) and PhD fellowship to Sughra Hakim by the Higher Education Commission (HEC) for research work is thankfully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 536 kb)
Rights and permissions
About this article
Cite this article
Hakim, S., Mirza, B.S., Zaheer, A. et al. Retrieved 16S rRNA and nifH sequences reveal co-dominance of Bradyrhizobium and Ensifer (Sinorhizobium) strains in field-collected root nodules of the promiscuous host Vigna radiata (L.) R. Wilczek. Appl Microbiol Biotechnol 102, 485–497 (2018). https://doi.org/10.1007/s00253-017-8609-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8609-6