Abstract
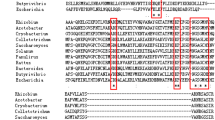
A glutamine synthetase (GS; 1341 bp) gene with potent L-phosphinothricin (PPT) resistance was isolated and characterized from a marine bacterium Exiguobacterium sp. Molecular docking analysis indicated that the substitution of residues Glu60 and Arg64 may lead to significant changes in binding pocket. To enhance the enzymatic property of GS, variants E60A and R64G were obtained by site-directed mutagenesis. The results revealed a noteworthy change in the thermostability and activity in comparison to the wild type (WT). WT exhibited optimum activity at 35 °C, while E60A and R64G exhibited optimum activity at 45 and 40 °C, respectively. The mutant R64G was 4.3 times more stable at 70 °C in comparison to WT, while E60A was 5.7 times more stable. Kinetic analysis revealed that the k cat value of R64G mutant was 8.10-, 7.25- and 7.63-fold that of WT for ADP, glutamine and hydroxylamine, respectively. The kinetic inhibition (K i, 4.91 ± 0.42 mM) of R64G was 2.02-fold that of WT (2.43 ± 0.14 mM) for L-phosphinothricin. The analysis of structure and function relationship showed that the binding pocket underwent dramatic changes when Arg site of 64 was substituted by Gly, thus promoting the rapid capture of substrates and leading to increase in activity and PPT-resistance of mutant R64G. The rearrangements of the residues at the molecular level formed new hydrogen bonds around the active site, which contributed to the increase of thermostability of enzymes. This study provides new insights into substrate binding mechanism of glutamine synthetase and the improved GS gene also has a potential for application in transgenic crops with L-phosphinothricin tolerance.


Similar content being viewed by others
References
Almassy RJ, Janson CA, Hamlin R, Xuong N, Eisenberg D (1986) Novel subunit—subunit interactions in the structure of glutamine synthetase. Nature 323(6086):304–309
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22(2):195–201
Bayer E, Gugel KH, Hagele K, Hagenmaier H, Jessipow S, Konig WA, Zahner H (1972) Metabolic products of microorganisms. 98. Phosphinothricin and phosphinothricyl-alanyl-analine. Helvetica chimica acta 55(1):224–239. doi:10.1002/hlca.19720550126
Bender RA, Janssen KA, Resnick AD, Blumenberg M, Foor F, Magasanik B (1977) Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J Bacteriol 129(2):1001–1009
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27(3):343–350. doi:10.1093/bioinformatics/btq662
Bindal S, Gupta R (2014) L-theanine synthesis using gamma-glutamyl transpeptidase from Bacillus licheniformis ER-15. J Agric Food Chem 62(37):9151–9159. doi:10.1021/jf5022913
Brown C, Macdonald-Brown DS, Meers J (1974) Physiological aspects of microbial inorganic nitrogen metabolism. Adv Microb Physiol 11:1–52
Brown JR, Masuchi Y, Robb FT, Doolittle WF (1994) Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J Mol Evol 38(6):566–576
Chanjuan L, Hong Y, Shao Z, Lin L, Huang X, Liu P, Wu G, Meng X, Liu Z (2009) Novel alkali-stable, cellulase-free xylanase from deep-sea Kocuria sp. Mn22. J Microbiol Biotechn 19(9):873–880
Chronopoulou EG, Labrou NE (2011) Site-saturation mutagenesis: a powerful tool for structure-based design of combinatorial mutation libraries. Curr Protoc Protein Sci . doi:10.1002/0471140864.ps2606s6326.6. 1-26.6. 10
Eisenberg D, Gill HS, Pfluegl GM, Rotstein SH (2000) Structure–function relationships of glutamine synthetases. BBA-Protein Struct M 1477(1):122–145
Erfle J, Sauer F, Mahadevan S (1977) Effect of ammonia concentration on activity of enzymes of ammonia assimilation and on synthesis of amino acids by mixed rumen bacteria in continuous culture. J Dairy Sci 60(7):1064–1072
Fraser AR, Ridley SM (1984) Kinetics for glutamine-synthetase inhibition by phosphinothricin and measurement of other enzyme activities in situ in isolated asparagus cells using a freeze-thaw technique. Planta 161(5):470–474
Gill HS, Eisenberg D (2001) The crystal structure of phosphinothricin in the active site of glutamine synthetase illuminates the mechanism of enzymatic inhibition. Biochemistry 40(7):1903–1912
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18(15):2714–2723. doi:10.1002/elps.1150181505
Hashimoto W, Suzuki H, Yamamoto K, Kumagai H (1995) Effect of site-directed mutations on processing and activity of γ-glutamyltranspeptidase of Escherichia coli K-12. J Biochem 118(1):75–80
Hoshida H, Tanaka Y, Hibino T, Hayashi Y, Tanaka A, Takabe T, Takabe T (2000) Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol Biol 43(1):103–111
Khan MIH, Ito K, Kim H, Ashida H, Ishikawa T, Shibata H, Sawa Y (2005) Molecular properties and enhancement of thermostability by random mutagenesis of glutamate dehydrogenase from Bacillus subtilis. Biosci Biotech Bioch 69(10):1861–1870
Kim JN, Cann IKO, Mackie RI (2012) Purification, characterization, and expression of multiple glutamine synthetases from Prevotella ruminicola 23. J Bacteriol 194(1):176–184. doi:10.1128/Jb.05916-11
Krajewski WW, Jones TA, Mowbray SL (2005) Structure of Mycobacterium tuberculosis glutamine synthetase in complex with a transition-state mimic provides functional insights. P Natl Acad Sci USA 102(30):10499–10504. doi:10.1073/pnas.0502248102
Krajewski WW, Collins R, Holmberg-Schiavone L, Jones TA, Karlberg T, Mowbray SL (2008) Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J Mol Biol 375(1):217–228
Mu W, Zhang T, Jiang B (2015) An overview of biological production of L-theanine. Biotechnol Adv. doi:10.1016/j.biotechadv.2015.04.004
Murray DS, Chinnam N, Tonthat NK, Whitfill T, Wray LV, Fisher SH, Schumacher MA (2013) Structures of the Bacillus subtilis glutamine synthetase dodecamer reveal large intersubunit catalytic conformational changes linked to aunique feedback inhibition mechanism. J Biol Chem 288(50):35801–35811. doi:10.1074/jbc.M113.519496
Nilsson MT, Krajewski WW, Yellagunda S, Prabhumurthy S, Chamarahally GN, Siddamadappa C, Srinivasa BR, Yahiaoui S, Larhed M, Karlén A (2009) Structural basis for the inhibition of Mycobacterium tuberculosis glutamine synthetase by novel ATP-competitive inhibitors. J Mol Biol 393(2):504–513
Packer MS, Liu DR (2015) Methods for the directed evolution of proteins. Nat Rev Genet 16(7):379–394
Pesole G, Gissi C, Lanave C, Saccone C (1995) Glutamine synthetase gene evolution in bacteria. Mol Biol Evol 12(2):189–197
Rhee S, Chock P, Wedler F, Sugiyama Y (1981) Subunit interaction in unadenylylated glutamine synthetase from Escherichia coli.: evidence from methionine sulfoximine inhibition studies. J Biol Chem 256(2):644–648
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234(3):779–815. doi:10.1006/jmbi.1993.1626
Seabra AR, Pereira PA, Becker JD, Carvalho HG (2012) Inhibition of glutamine synthetase by phosphinothricin leads to transcriptome reprograming in root nodules of Medicago truncatula. Mol Plant Microbe In: MPMI 25(7):976–992. doi:10.1094/MPMI-12-11-0322
Sun H, Huang QM, Su J (2005) Highly effective expression of glutamine synthetase genes GS1 and GS2 in transgenic rice plants increases nitrogen-deficiency tolerance. J Plant Physiol Mol Biol 31(5):492–498
Tardito S, Oudin A, Ahmed SU, Fack F, Keunen O, Zheng L, Miletic H, Sakariassen PO, Weinstock A, Wagner A, Lindsay SL, Hock AK, Barnett SC, Ruppin E, Morkve SH, Lund-Johansen M, Chalmers AJ, Bjerkvig R, Niclou SP, Gottlieb E (2015) Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nat Cell Biol 17(12):1556–1568. doi:10.1038/ncb3272
Tian YS, Xu J, Zhao W, Xing XJ, Fu XY, Peng RH, Yao QH (2015) Identification of a phosphinothricin-resistant mutant of rice glutamine synthetase using DNA shuffling. Sci Rep-UK 5(15495). doi:10.1038/srep15495
Unno H, Uchida T, Sugawara H, Kurisu G, Sugiyama T, Yamaya T, Sakakibara H, Hase T, Kusunoki M (2006) Atomic structure of plant glutamine synthetase: a key enzyme for plant productivity. J Biol Chem 281(39):29287–29296
Van Rooyen JM, Abratt VR, Belrhali H, Sewell T (2011) Crystal structure of type III glutamine synthetase: surprising reversal of the inter-ring interface. Structure 19(4):471–483
Vishnivetskaya TA, Lucas S, Copeland A, Lapidus A, del Rio TG, Dalin E, Tice H, Bruce DC, Goodwin LA, Pitluck S (2011) Complete genome sequence of the thermophilic Exiguobacterium sp. AT1b. J Bacteriol 193(11):2880–2881
Wang M, Si T, Zhao H (2012) Biocatalyst development by directed evolution. Bioresource Technol 115:117–125
Wedler F, Boyer P (1972) Substrate binding and reaction intermediates of glutamine synthetase (Escherichia coli W) as studied by isotope exchanges. J Biol Chem 247(4):984–992
Wray LV, Fisher SH (2008) Bacillus subtilis GlnR contains an autoinhibitory C-terminal domain required for the interaction with glutamine synthetase. Mol Microbiol 68(2):277–285
Wray LV, Fisher SH (2010) Functional roles of the conserved Glu304 loop of Bacillus subtilis glutamine synthetase. J Bacteriol 192(19):5018–5025
Yamamoto S, Wakayama M, Tachiki T (2006) Cloning and expression of Pseudomonas taetrolens Y-30 gene encoding glutamine synthetase: an enzyme available for theanine production by coupled fermentation with energy transfer. Biosci Biotech Bioch 70(2):500–507. doi:10.1271/Bbb.70.500
Zhou X, Zhang ZP, Jia XH, Wu YF, Luo L, Yin ZM (2008) Mn2+ enhances theanine-forming activity of recombinant glutamine synthetase from Bacillus subtilis in Escherichia coli. World J Microb Biot 24(8):1267–1272. doi:10.1007/s11274-007-9599-9
Acknowledgements
This work was supported by grants from the Genetically Modified Organisms Breeding Major Projects of China (2016ZX08001001001-001-001-004).
Author contribution statement
SWZ designed and performed the experiments and drafted this manuscript. YKH, AK and HG contributed to characterization, expression experiments and revision of the manuscript. NH and ZDL are corresponding authors, who conceived, designed and supervised the experiments. All authors have read and approved the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Declarations of interest
The authors claim that they have no conflict of interest in either writing the paper or in the project.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 1963 kb).
Rights and permissions
About this article
Cite this article
Zhang, S., Han, Y., Kumar, A. et al. Characterization of an L-phosphinothricin resistant glutamine synthetase from Exiguobacterium sp. and its improvement. Appl Microbiol Biotechnol 101, 3653–3661 (2017). https://doi.org/10.1007/s00253-017-8103-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8103-1