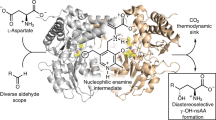
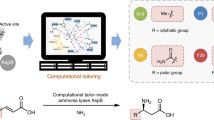
Abstract
Bacterial arylmalonate decarboxylase (AMDase) shows high enantioselectivity and a broad substrate spectrum in the asymmetric synthesis of optically pure arylaliphatic carboxylic acids. The determination of the structure of AMDase has greatly extended the understanding of the catalytic mechanism of this unique cofactor-free decarboxylase and allowed the generation of tailor-made enzyme variants with improved catalytic properties. Despite this increase in knowledge and applicability, the natural role of the enzyme remains unknown. This mini-review summarizes the recent findings on the molecular mechanism and the synthetic application of the enzyme.









Similar content being viewed by others
References
Appleby TC, Kinsland C, Begley TP, Ealick SE (2010) The crystal structure and mechanism of orotidine 5 -monophosphate decarboxylase. Proc Natl Acad Sci USA 97(5):2005–2010
Bornscheuer U, Huisman G, Kazlauskas R, Lutz S, Moore J, Robins K (2012) Engineering the third wave of biocatalysis. Nature 485(7397):185–194
Brück T, Kourist R, Loll B (2014) Production of macrocyclic sesqui-and diterpenes in heterologous microbial hosts: a systems approach to harness Nature’s molecular diversity. ChemCatChem 6(5):1142–1165
Busch F, Hülsemann N, Enoki J, Miyamoto K, Bocola M, Kourist R (2016) Semiempirical QM/MM calculations reveal a step-wise proton transfer and an unusual thiolate pocket in the mechanism of the racemizing mutant G74C of arylmalonate decarboxylase. Cat Sci Technol 6(13):4937–4944
Engström K, Nyhlen J, Sandström AG, Backväll JE (2010) Directed Evolution of an Enantioselective Lipase with Broad Substrate Scope for Hydrolysis of alpha-Substituted Esters. J Am Chem Soc 132(20):7038–7042
Fisch F, Fleites CM, Delenne M, Baudendistel N, Hauer B, Turkenburg JP, Hart S, Bruce NC, Grogan G (2010) A covalent succinylcysteine-like intermediate in the enzyme-catalyzed transformation of maleate to fumarate by maleate isomerase. J Am Chem Soc 132(33):11455–11457
Frank A, Eborall W, Hyde R, Hart S, Turkenburg JP, Grogan G (2012) Mutational analysis of phenolic acid decarboxylase from Bacillus subtilis (BsPAD), which converts bio-derived phenolic acids to styrene derivatives. Cat Sci Technol 2(8):1568–1574
Gaßmeyer SK, Yoshikawa H, Enoki J, Hülsemann N, Stoll R, Miyamoto K, Kourist R (2015) STD-NMR-based protein engineering of the unique arylpropionate-racemase AMDase G74C. ChemBioChem 16(13):1943–1949
Gaßmeyer S, Wetzig J, Mügge C, Assmann M, Enoki J, Hilterhaus L, Zuhse R, Miyamoto K, Liese A, Kourist R (2016) Arylmalonate decarboxylase-catalyzed asymmetric synthesis of both enantiomers of optically pure flurbiprofen. ChemCatChem 8:916–921
Giacomini D, Galletti P, Quintavalla A, Gucciardo G, Paradisi F (2007) Highly efficient asymmetric reduction of arylpropionic aldehydes by Horse Liver Alcohol Dehydrogenase through dynamic kinetic resolution. Chem Commun (39):4038–4040
Glueck SM, Gümüs S, Fabian WM, Faber K (2010) Biocatalytic carboxylation. Chem Soc Rev 39(1):313–328
Ijima, Y., K. Matoishi, Y. Terao, N. Doi, H. Yanagawa and H. Ohta (2005). Inversion of enantioselectivity of asymmetric biocatalytic decarboxylation by site-directed mutagenesis based on the reaction mechanism. Chem Commun 877–879
Kalb D, Gressler J, Hoffmeister D (2015) Active-site engineering expands the substrate profile of the basidiomycete L-tryptophan decarboxylase CsTDC. ChemBioChem 17:132–136
Korth M (2010) Third-generation hydrogen-bonding corrections for semiempirical QM methods and force fields. J Chem Th Comp 6(12):3808–3816
Könst P, Merkens H, Kara S, Kochius S, Vogel A, Zuhse R, Holtmann D, Arends IW, Hollmann F (2012) Enantioselective oxidation of aldehydes catalyzed by alcohol dehydrogenase. Angew Chem Int Ed 51(39):9914–9917
Kourist R, Miyamoto K (2011) Protein engineering of arylmalonate decarboxylase G74C, an artificial racemase. Biotechnol Intern 23:6–9
Kourist R, Miyauchi Y, Uemura D, Miyamoto K (2011) Engineering the promiscuous racemase activity of arylmalonate decarboxylase. Chem Eur J 17:557–563
Wong LS, Okrasa K, Micklefield J (2010) Site-selective immobilisation of functional enzymes on to polystyrene nanoparticles. Org Biomol Chem 8(4):782–787
Kourist R, Guterl J-K, Miyamoto K, Sieber V (2014) Enzymatic decarboxylation—an emerging reaction for chemicals production from renewable resources. ChemCatChem 6(3):689–701
Kuettner EB, Keim A, Kircher M, Rosmus S, Strater N (2008) Active-site mobility revealed by the crystal structure of arylmalonate decarboxylase from Bordetella bronchiseptica. J Mol Biol 377(2):386–394
Lewin R, Goodall M, Thompson ML, Leigh J, Breuer M, Baldenius K, Micklefield J (2015) Enzymatic enantioselective decarboxylative protonation of heteroaryl malonates. Chem Eur J 21(17):6557–6563
Lind ME, Himo F (2014) Theoretical study of reaction mechanism and stereoselectivity of arylmalonate decarboxylase. ACS Catal 4(11):4153–4160
Maimanakos J, Chow J, Gaßmeyer S, Kourist S, Streit WR (2016) Sequence-based screening for rare enzymes: new insights into the world of AMDases. Front Microbiol. doi:10.3389/fmicb.2016.01332
Matoishi K, Ueda M, Miyamoto K, Ohta H (2004) Mechanism of asymmetric decarboxylation of α-aryl-α-methylmalonate catalyzed by arylmalonate decarboxylase originated from Alcaligenes bronchisepticus. J Mol Catal B Enzym 27(4):161–168
May M, Mehboob S, Mulhearn DC, Wang Z, Yu H, Thatcher GRJ, Santarsiero BD, Johnson ME, Mesecar AD (2007) Structural and functional analysis of two glutamate racemase isozymes from Bacillus anthracis and implications for inhibitor design. J Mol Biol 371(5):1219–1237
Miyamoto K, Ohta H (1990) Enzyme-mediated asymmetric decarboxylation of disubstituted malonic acids. J Am Chem Soc 112:4077–4078
Miyamoto K, Ohta H (1992a) Cloning and heterologous expression of a novel arylmalonate decarboxylase gene from Alcaligenes bronchisepticus KU 1201. Appl Microbiol Biotechnol 38(2):234–238
Miyamoto K, Ohta H (1992b) Purification and properties of a novel arylmalonate decarboxylase from Alcaligenes bronchosepticus KU-1201. Eur J Biochem 210(2):475–481
Miyamoto K, Tsuchiya S, Ohta H (1992) Stereochemistry of enzyme-catalyzed decarboxylation of alpha-methyl-alpha-phenylmalonic acid. J Am Chem Soc 2:6256–6257
Miyamoto K, Tsutsumi T, Terao Y, Ohta H (2007a) Stereochemistry of decarboxylation of arylmalonate catalyzed by mutant enzymes. Chem Lett 36(5):656–657
Miyamoto K, Yatake Y, Tamura K, Terao Y, Ohta H (2007b) Purification and characterization of arylmalonate decarboxylase from Achromobacter sp KU1311. J Biosc Bioeng 104(4):263–267
Miyauchi Y, Kourist R, Uemura D, Miyamoto K (2011) Dramatically improved catalytic activity of an artificial (S)-selective arylmalonate decarboxylase by structure-guided directed evolution. Chem Commun 47:7503–7505
Obata R, Nakasako M (2010) Structural basis for inverting the enantioselectivity of arylmalonate decarboxylase revealed by the structural analysis of the Gly74Cys/Cys188Ser mutant in the liganded form. Biochemistry 49:1963–1969
Okrasa K, Levy C, Hauer B, Baudendistel N, Leys D, Micklefield J (2008) Structure and mechanism of an unusual malonate decarboxylase and related racemases. Chem Eur J 14(22):6609–6613
Okrasa K, Levy C, Wilding M, Goodall M, Baudendistel N, Hauer B, Leys D, Micklefield J (2009) Structure-guided directed evolution of alkenyl and arylmalonate decarboxylases. Angew Chem Int Ed 48(41):7691–7694
Payne KA, White MD, Fisher K, Khara B, Bailey SS, Parker D, Rattray NJ, Trivedi DK, Goodacre R, Beveridge R (2015) New cofactor supports [agr],[bgr]-unsaturated acid decarboxylation via 1, 3-dipolar cycloaddition. Nature 522(7557):497–501
Puig E, Garcia-Viloca M, González-Lafont À, Lluch JM (2006) On the ionization state of the substrate in the active site of glutamate racemase. A QM/MM study about the importance of being zwitterionic. J Phys Chem A 110(2):717–725
Puig E, Mixcoha E, Garcia-Viloca M, Gonzalez-Lafont A, Lluch JM (2009) How the substrate D-glutamate drives the catalytic action of Bacillus subtilis glutamate racemase. J Am Chem Soc 131(10):3509–3521
Sandström AG, Wikmark Y, Engström K, Nyhlén J, J-e B (2012) Combinatorial reshaping of the Candida antarctica lipase A substrate pocket for enantioselectivity using an extremely condensed library. Proc Natl Acad Sci U S A 109:78–83
Steenkamp L, Brady D (2003) Screening of commercial enzymes for the enantio selective hydrolysis of R, S-naproxen ester. Enzym Microb Technol 32(3–4):472–477
Steenkamp L, Brady D (2008) Optimisation of stabilised Carboxylesterase NP for enantioselective hydrolysis of naproxen methyl ester. Process Biochem 43:1419–1426
Strübing D, Krumlinde P, Piera J, Bäckvall JE (2007) Dynamic kinetic resolution of primary alcohols with an unfunctionalized stereogenic center in the beta-position. Adv Synth Catal 349:1577–1581
Tamura K, Terao Y, Miyamoto K, Ohta H (2008) Asymmetric decarboxylation of alpha-hydroxy- and alpha-amino-alpha-phenylmalonate catalyzed by arylmalonate decarboxylase from Alcaligenes bronchisepticus. Biocatal Biotransform 26(4):253–257
Terao Y, Ijima Y, Kakidani H, Ohta H (2003) Enzymatic synthesis of (R)-flurbiprofen. Bull Chem Soc Japan 76(12):2395–2397
Terao Y, Miyamoto K, Ohta H (2006a) Improvement of the activity of arylmalonate decarboxylase by random mutagenesis. Appl Microbiol Biotechnol 73(3):647–653
Terao Y, Miyamoto K, Ohta H (2006b) Introduction of single mutation changes arylmalonate decarboxylase to racemase. Chem Commun 34:3600–3602
Um PJ, Drueckhammer DG (1998) Dynamic enzymatic resolution of thioesters. J Am Chem Soc 120(23):5605–5610
Vardi-Kilshtain A, Doron D, Major DT (2013) Quantum and classical simulations of orotidine monophosphate decarboxylase: support for a direct decarboxylation mechanism. Biochemistry 52(25):4382–4390
Yatake Y, Miyamoto K, Ohta H (2008) Screening, cloning, expression, and purification of an acidic arylmalonate decarboxylase from Enterobacter cloacae KU1313. Appl Microbiol Biotechnol 78(5):793–799
Yoshida S, Enoki J, Hemmi R, Kourist R, Kawakami N, Miyamoto K (2015a) Draft genome sequence of Bordetella bronchiseptica KU1201, the first isolation source of arylmalonate decarboxylase. Genome Ann 3(3):e00373–e00315
Yoshida S, Enoki J, Kourist R, Miyamoto K (2015b) Engineered hydrophobic pocket of (S)-selective arylmalonate decarboxylase variant by simultaneous saturation mutagenesis to improve catalytic performance. Biosc Biotechnol Biochem 79:1965–1971
Zachos I, Gaßmeyer SK, Bauer D, Sieber V, Hollmann F, Kourist R (2015) Photobiocatalytic decarboxylation for olefin synthesis. Chem Commun 51:1918–1921
Acknowledgments
The authors thank Florian Busch (Ruhr-Universität Bochum, Germany) and Marco Bocola (RWTH Aachen, Germany) for very insightful suggestions regarding the mechanism of AMDase and Janine Maimanakos for discussions of the biological distribution of the enzyme and the phylogenetic analysis of the asp/glu racemase superfamily. The authors thank the North Rhine-Westphalian Ministry for Innovation, Science and Investigation (award number PtJ-TRI/1411ng006) and the German Academic Exchange Service (award number 57154401) for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the North Rhine-Westphalian Ministry for Innovation, Science and Investigation (award number PtJ-TRI/1411ng006) and the German Academic Exchange Service (award number 57154401).
Conflict of interest
Robert Kourist declares that he has no conflict of interest.
Kenji Miyamoto declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.”
Rights and permissions
About this article
Cite this article
Miyamoto, K., Kourist, R. Arylmalonate decarboxylase—a highly selective bacterial biocatalyst with unknown function. Appl Microbiol Biotechnol 100, 8621–8631 (2016). https://doi.org/10.1007/s00253-016-7778-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7778-z