Abstract
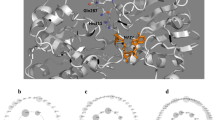
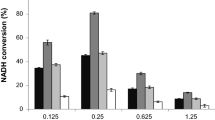
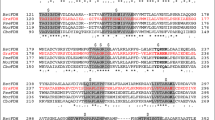
Formate dehydrogenases (FDHs) are continually used for the cofactor regeneration in biocatalysis and biotransformation with hiring NAD(P)H-dependent oxidoreductases. Major weaknesses of most native FDHs are their low activity and operational stability in the catalytic reaction. In this work, the FDH from Candida boidinii (CboFDH) was engineered in order to gain an enzyme with high activity and better operational stability. Through comparing and analyzing its spatial structure with other FDHs, the catalysis, substrate, and coenzyme binding sites of the CboFDH were identified. To improve its performance, amino acids, which concentrated on the enzyme active site or in the conserved NAD+ and substrate binding motif, were mutated. The mutant V120S had the highest catalytic efficiency (k cat/K m ) with COONH4 as it enhanced the catalytic velocity (k cat) and k cat/K m 3.48-fold and 1.60-fold, respectively, than that of the wild type. And, the double-mutant V120S-N187D had the highest k cat/K m with NAD+ as it displayed an approximately 1.50-fold increase in k cat/K m . The mutants showed higher catalytic efficiency than other reported FDHs, suggesting that the mutation has achieved good results. The single and double mutants exhibited higher thermostability than the wild type. The structure-function relationship of single and double mutants was analyzed by homology models and site parsing. Asymmetric synthesis of L-tert-leucine was executed to evaluate the ability of cofactor regeneration of the mutants with about 100 % conversion rates. This work provides a helpful theoretical reference for the evolution of an enzyme in vitro and promotion of the industrial production of chiral compounds, e.g., amino acid and chiral amine.








Similar content being viewed by others
References
Addo PK, Arechederra RL, Minteer S (2010) Evaluating enzyme cascades for methanol/air biofuel cells based on NAD+-dependent enzymes. Electroanalysis 22(7–8):807–812
Akers NL, Moore CM, Minteer SD (2005) Development of alcohol/O2 biofuel cells using salt-extracted tetrabutylammonium bromide/Nafion membranes to immobilize dehydrogenase enzymes. Electrochim Acta 50(12):2521–2525
Ansorge-Schumacher MB, Slusarczyk H, Schümers J, Hirtz D (2006) Directed evolution of formate dehydrogenase from Candida boidinii for improved stability during entrapment in polyacrylamide. FEBS J 273(17):3938–3945
Baik SH, Ide T, Yoshida H, Kagami O, Harayama S (2003) Significantly enhanced stability of glucose dehydrogenase by directed evolution. Appl Microbiol Biotechnol 61(4):329–335
Bhardwaj A, Leelavathi S, Mazumdar-Leighton S, Ghosh A, Ramakumar S, Reddy VS (2010) The critical role of N- and C-terminal contact in protein stability and folding of a family 10 xylanase under extreme conditions. PLoS One 5(6):3039–3056
Bolivar JM, Lorena W, Susana Alicia F, Roberto FL, Guisan JM, Cesar M (2006) Stabilization of a formate dehydrogenase by covalent immobilization on highly activated glyoxyl-agarose supports. Biomacromolecules 7(3):669–673
Carter JL, Bekhouche M, Noiriel A, Blum LJ, Doumèche B (2014) Directed evolution of a formate dehydrogenase for increased tolerance to ionic liquids reveals a new site for increasing the stability. ChemBioChem
Demir AS, Talpur FN, Betul Sopaci S, Kohring GW, Ayhan C (2011) Selective oxidation and reduction reactions with cofactor regeneration mediated by galactitol-, lactate-, and formate dehydrogenases immobilized on magnetic nanoparticles. J Biotechnol 152(4):176–183
Donk WA, Der V, Huimin Z (2003) Recent developments in pyridine nucleotide regeneration. Curr Opin Biotechnol 14(4):421–426
Filippova EV, Polyakov KM, Tikhonova TV, Stekhanova TN, Boiko KM, Popov VO (2005) Structure of a new crystal modification of the bacterial NAD-dependent formate dehydrogenase with a resolution of 2.1 Å. Crystallogr Rep 50(5):796–800
Fischer T, Pietruszka J (2010) Key building blocks via enzyme-mediated synthesis. Springer, Berlin
Fogal S, Beneventi E, Cendron L, Bergantino E (2015) Structural basis for double cofactor specificity in a new formate dehydrogenase from the acidobacterium Granulicella mallensis MP5ACTX8. Appl Microbiol Biotechnol 1–14
Funahashi J, Takano K, Yamagata Y, Yutani K (2000) Role of surface hydrophobic residues in the conformational stability of human lysozyme at three different positions. Biochemistry 39(47):14448–14456
Gul-Karaguler N, Sessions RB, Clarke AR, Holbrook JJ (2001) A single mutation in the NAD-specific formate dehydrogenase from Candida methylica allows the enzyme to use NADP. Biotechnol Lett 23(4):283–287
Hatrongjit R, Packdibamrung K (2010) A novel NADP+-dependent formate dehydrogenase from Burkholderia stabilis 15516: screening, purification and characterization. Enzym Microb Technol 46(7):557–561
Hoelsch K, Sührer I, Heusel M, Weuster-Botz D (2012) Engineering of formate dehydrogenase: synergistic effect of mutations affecting cofactor specificity and chemical stability. Appl Microbiol Biotechnol 97(6):2473–2481
Hong W, Huang S, Jiang Z (2004) Effects of modification of silica gel and ADH on enzyme activity for enzymatic conversion of CO2 to methanol. Catal Today 98(4):545–552
Jeon JK, Ihm SK, Park YK, Kim JS, Dong JI, Kim S, Kim JM, Kim SS, Yoo KS (2004) Membrane/PSA hybrid process for carbon dioxide recovery at low concentration. Stud Surf Sci Catal 153(04):543–546
Jiang W, Chen L, Yuan S, Li B, Liu Z (2014) A novel serine hydroxymethyltransferase from Arthrobacter nicotianae: characterization and improving catalytic efficiency by rational design. BMC Biotechnol 14(1):93
Jiang W, Sun D, Lu J, Wang Y, Wang S, Zhang Y, Fang B (2015) A cold-adapted leucine dehydrogenase from marine bacterium Alcanivorax dieselolei: characterization and L-tert -leucine production. Eng Life Sci 12(11)
Koutinas AA, Vlysidis A, Pleissner D, Kopsahelis N, Garcia IL, Kookos IK, Papanikolaou S, Kwan TH, Lin CSK (2014) Valorization of industrial waste and by-product streams via fermentation for the production of chemicals and biopolymers. Chem Soc Rev 43(8):2587–2627
Kula M-R, Pohl M, Slusarczyk H (2003) Mutanten der Formiatdehydrogenase aus Candida biodinii, Gensequenzen diese codierend sowie Verwendung dieser Formiatdeydrogenasen. EP
Lu Y, Jiang ZY, Xu SW, Wu H (2006) Efficient conversion of CO2 to formic acid by formate dehydrogenase immobilized in a novel alginate–silica hybrid gel. Catal Today 115(s 1–4):263–268
Mourad B, Blum LCJ, Bastien D (2011) Contribution of dynamic and static quenchers for the study of protein conformation in ionic liquids by steady-state fluorescence spectroscopy. J Phys Chem B 116(1):413–423
Munro PA, Dunnill P, Lilly MD (1975) Magnetic particles as supports in immobilised enzyme reactors. Magn IEEE Trans 11(5):1573–1575
Netto CGCM, Nakamura M, Andrade LH, Toma HE (2012) Improving the catalytic activity of formate dehydrogenase from Candida boidinii by using magnetic nanoparticles. J Mol Catal B Enzym 84(4):136–143
Nilov DK, Shabalin IG, Popov VO, Svedas VK (2012) Molecular modeling of formate dehydrogenase: the formation of the Michaelis complex. J Biomol Struct Dyn 30(2):170–179
Perl D, Mueller U, Heinemann U, Schmid FX (2000) Two exposed amino acid residues confer thermostability on a cold shock protein. Nat Struct Mol Biol 7(5):380–383
Popov VO, Lamzin VS (1994) NAD+-dependent formate dehydrogenase. Biochem J 301((Pt 3)(3)):625–643
Schirwitz K, Schmidt A, Lamzin VS (2007) High-resolution structures of formate dehydrogenase from Candida boidinii. Protein Sci 16(6):1146–1156
Serov A, Popova A, Fedorchuk V, Tishkov V (2002) Engineering of coenzyme specificity of formate dehydrogenase from Saccharomyces cerevisiae. Biochem J 367:841–847
Shaked Z, Whitesides GM (1980) Enzyme-catalyzed organic synthesis: NADH regeneration by using formate dehydrogenase. J Am Chem Soc 102(23):7104–7105
Slusarczyk H, Felber S, Kula MR, Pohl M (2000) Stabilization of NAD-dependent formate dehydrogenase from Candida boidinii by site-directed mutagenesis of cysteine residues. Eur J Biochem 267(5):1280–1289
Tishkov VI, Popov VO (2006) Protein engineering of formate dehydrogenase. Biomol Eng 23(2):89–110
Wichmann R, Vasic-Racki D (2005) Cofactor regeneration at the lab scale. Adv Biochem Eng/Biotechnol 92:225–260
Wohlgemuth R (2010) Asymmetric biocatalysis with microbial enzymes and cells. Curr Opin Microbiol 13(3):283–292
Xu HQY, Huang Z, Liu Z (2014) Characterization and site-directed mutagenesis of α-galactosidase from the deep-sea bacterium Bacillus megaterium. Enzym Microb Technol 56:46–52
Yoshimoto M, Yamasaki R, Nakao M, Yamashita T (2010) Stabilization of formate dehydrogenase from Candida boidinii through liposome-assisted complexation with cofactors. Enzym Microb Technol 46(7):588–593
Acknowledgments
This work was supported by the State Key Program of National Natural Science Foundation of China (No. 21336009), the National Natural Science Foundation of China (Nos. 41176111 and 41306124), the Fundamental Research Funds for the Central Universities (No. 2013121029), the Foundation of South Oceanographic Research Center of China in Xiamen (No. 14GYY011NF11), and the Public Science and Technology Research Funds Projects of Ocean (No. 201505032–6).
Authors’ contributions
WJ directed the research, performed the experiment, and wrote the manuscript. PL and RNY assisted with the steady-state kinetic study. BSF conceived the project, directed the research, and supervised the work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors state that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 994 kb)
Rights and permissions
About this article
Cite this article
Jiang, W., Lin, P., Yang, R. et al. Identification of catalysis, substrate, and coenzyme binding sites and improvement catalytic efficiency of formate dehydrogenase from Candida boidinii . Appl Microbiol Biotechnol 100, 8425–8437 (2016). https://doi.org/10.1007/s00253-016-7613-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-7613-6