Abstract
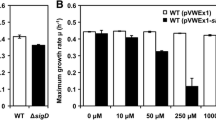
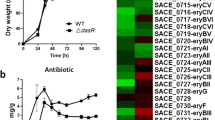
The roles of many sigma factors are unclear in regulatory mechanism of secondary metabolism in Streptomyces. Here, we report the regulation network of a group 3 sigma factor, WhiGch, from a natamycin industrial strain Streptomyces chattanoogensis L10. WhiGch regulates the growth and morphological differentiation of S. chattanoogensis L10. The whiG ch deletion mutant decreased natamycin production by about 30 % and delayed natamycin production more than 24 h by delaying the growth. Overexpression of the whiG ch gene increased natamycin production in large scale production medium by about 26 %. WhiGch upregulated the transcription of natamycin biosynthetic gene cluster and inhibited the expression of migrastatin and jadomycin analog biosynthetic polyketide synthase genes. WhiGch positively regulated natamycin biosynthetic gene cluster by directly binding to the promoters of scnC and scnD, which were involved in natamycin biosynthesis, and these binding sites adjacent to translation start codon were determined. Thus, this paper further elucidates the high natamycin yield mechanisms of industrial strains and demonstrates that a valuable improvement in the yield of the target metabolites can be achieved through manipulating the transcription regulators.






Similar content being viewed by others
References
Ainsa JA, Parry HD, Chater KF (1999) A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol Microbiol 34(3):607–619. doi:10.1046/j.1365-2958.1999.01630.x
Catakli S, Andrieux A, Decaris B, Dary A (2005) Sigma factor WhiG and its regulation constitute a target of a mutational phenomenon occurring during aerial mycelium growth in Streptomyces ambofaciens ATCC23877. Res Microbiol 156(3):328–340. doi:10.1016/j.resmic.2004.12.001
Chater KF (1972) A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J Gen Microbiol 72(1):9–28
Chater KF (1975) Construction and phenotypes of double sporulation deficient mutants in Streptomyces coelicolor A3(2). J Gen Microbiol 87(2):312–325
Chater KF (2001) Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr Opin Microbiol 4(6):667–673
Chater KF, Bruton CJ, Plaskitt KA, Buttner MJ, Mendez C, Helmann JD (1989) The developmental fate of S.coelicolor hyphae depends upon a gene product homologous with the motility sigma factor of B. subtilis. Cell 59(1):133–143. doi:10.1016/0092-8674(89)90876-3
Chen GQ, Lu FP, Du LX (2008) Natamycin production by Streptomyces gilvosporeus based on statistical optimization. J Agric Food Chem 56(13):5057–5061. doi:10.1021/jf800479u
Cho YH, Lee EJ, Ahn BE, Roe JH (2001) SigB, an RNA polymerase sigma factor required for osmoprotection and proper differentiation of Streptomyces coelicolor. Mol Microbiol 42(1):205–214
Du YL, Chen SF, Cheng LY, Shen XL, Tian Y, Li YQ (2009) Identification of a novel Streptomyces chattanoogensis L10 and enhancing its natamycin production by overexpressing positive regulator ScnRII. J Microbiol 47(4):506–513. doi:10.1007/s12275-009-0014-0
Du YL, Li SZ, Zhou Z, Chen SF, Fan WM, Li YQ (2011a) The pleitropic regulator AdpAch is required for natamycin biosynthesis and morphological differentiation in Streptomyces chattanoogensis. Microbiol-SGM 157:1300–1311. doi:10.1099/Mic.0.046607-0
Du YL, Shen XL, Yu P, Bai LQ, Li YQ (2011b) γ-butyrolactone regulatory system of Streptomyces chattanoogensis links nutrient utilization, metabolism, and development. Appl Environ Microbiol 77(23):8415–8426. doi:10.1128/Aem.05898-11
Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30(1):207–210
el-Enshasy HA, Farid MA, el-Sayed SA (2000) Influence of inoculum type and cultivation conditions on natamycin production by Streptomyces natalensis. J Basic Microbiol 40(5–6):333–342
Fink JS, Verhave M, Kasper S, Tsukada T, Mandel G, Goodman RH (1988) The CGTCA sequence motif is essential for biological activity of the vasoactive intestinal peptide gene cAMP-regulated enhancer. Proc Natl Acad Sci U S A 85(18):6662–6666
Genay M, Catakli S, Kleinclauss A, Andrieux A, Decaris B, Dary A (2006) Genetic instuability of whiG gene during the aerial mycelium development of Streptomyces ambofaciens ATCC23877 under different conditions of nitrogen limitations. Mutat Res Fundam Mol Mech 595(1–2):80–90
Genay M, Decaris B, Dary A (2007) Implication of stringent response in the increase of mutability of the whiG and whiH genes during Streptomyces coelicolor development. Mutat Res Fundam Mol Mech 624(1–2):49–60. doi:10.1016/j.mrfmmm.2007.03.016
Gomez-Santos N, Perez J, Sanchez-Sutil MC, Moraleda-Munoz A, Munoz-Dorado J (2011) CorE from Myxococcus xanthus is a copper-dependent RNA polymerase sigma factor. PLoS Genet 7(6):e1002106. doi:10.1371/journal.pgen.1002106
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100(4):1541–1546. doi:10.1073/pnas.0337542100
Han L, Yang K, Ramalingam E, Mosher RH, Vining LC (1994) Cloning and characterization of polyketide synthase genes for jadomycin B biosynthesis in Streptomyces venezuelae ISP5230. Microbiol-SGM 140(12):3379–3389
Helmann JD (1991) Alternative sigma factors and the regulation of flagellar gene-expression. Mol Microbiol 5(12):2875–2882. doi:10.1111/j.1365-2958.1991.tb01847.x
Helmann JD (2002) The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 46:47–110
Ishihama A (2000) Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol 54:499–518. doi:10.1146/annurev.micro.54.1.499
Jiang H, Wang YY, Ran XX, Fan WM, Jiang XH, Guan WJ, Li YQ (2013) Improvement of natamycin production by engineering of phosphopantetheinyl transferases in Streptomyces chattanoogensis L10. Appl Environ Microbiol 79(11):3346–3354. doi:10.1128/AEM.00099-13
Kieser TBM, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. The John Innes Foundation, Norwich
Lim SK, Ju J, Zazopoulos E, Jiang H, Seo JW, Chen Y, Feng Z, Rajski SR, Farnet CM, Shen B (2009) iso-Migrastatin, migrastatin, and dorrigocin production in Streptomyces platensis NRRL 18993 is governed by a single biosynthetic machinery featuring an acyltransferase-less type I polyketide synthase. J Biol Chem 284(43):29746–29756. doi:10.1074/jbc.M109.046805
Liu J, Li J, Wu Z, Pei H, Zhou J, Xiang H (2012) Identification and characterization of the cognate anti-sigma factor and specific promoter elements of a T. tengcongensis ECF sigma factor. PLoS ONE 7(7):e40885. doi:10.1371/journal.pone.0040885
Lu CG, Liu WC, Qiu JY, Wang HM, Liu T, Liu DW (2008) Identification of an antifungal metabolite produced by a potential biocontrol actinomyces strain A01. Braz J Microbiol 39(4):701–707
Mao XM, Zhou Z, Cheng LY, Hou XP, Guan WJ, Li YQ (2009a) Involvement of SigT and RstA in the differentiation of Streptomyces coelicolor. FEBS Lett 583(19):3145–3150. doi:10.1016/j.febslet.2009.09.025
Mao XM, Zhou Z, Hou XP, Guan WJ, Li YQ (2009b) Reciprocal regulation between SigK and differentiation programs in Streptomyces coelicolor. J Bacteriol 191(21):6473–6481. doi:10.1128/JB.00875-09
Mao XM, Sun N, Wang F, Luo S, Zhou Z, Feng WH, Huang FL, Li YQ (2013) Dual positive feedback regulation of protein degradation of an extra cytoplasmic function sigma factor for cell differentiation in Streptomyces coelicolor. J Biol Chem 288(43):31217–31228. doi:10.1074/jbc.M113.491498
Mendez C, Chater KF (1987) Cloning of whiG, a gene critical for sporulation of Streptomyces coelicolor A3(2). J Bacteriol 169(12):5715–5720
Ryding NJ, Kelemen GH, Whatling CA, Flardh K, Buttner MJ, Chater KF (1998) A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol Microbiol 29(1):343–357. doi:10.1046/j.1365-2958.1998.00939.x
Sambrook JFE, Maniatis T (2000) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Santos-Aberturas J, Vicente CM, Guerra SM, Payero TD, Martin JF, Aparicio JF (2011) Molecular control of polyene macrolide biosynthesis direct binding of the regulator pimM to eight promoters of pimaricin genes and identification of binding boxes. J Biol Chem 286(11):9150–9161. doi:10.1074/jbc.M110.182428
Sun J, Kelemen GH, Fernandez-Abalos JM, Bibb MJ (1999) Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiol-SGM 145(9):2221–2227
Tan HR, Yang HH, Tian YQ, Wu W, Whatling CA, Chamberlin LC, Buttner MJ, Nodwell J, Chater KF (1998) The Streptomyces coelicolor sporulation-specific sigma (WhiG) form of RNA polymerase transcribes a gene encoding a ProX-like protein that is dispensable for sporulation. Gene 212(1):137–146. doi:10.1016/S0378-1119(98)00152-8
Wang L, Vining LC (2003) Control of growth, secondary metabolism and sporulation in Streptomyces venezuelae ISP5230 by jadW1, a member of the afsA family of γ-butyrolactone regulatory genes. Microbiol-SGM 149(8):1991–2004
Wang G, Tanaka Y, Ochi K (2010) The G243D mutation (afsB mutation) in the principal sigma factor sigma HrdB alters intracellular ppGpp level and antibiotic production in Streptomyces coelicolor A3(2). Microbiology 156(Pt 8):2384–2392. doi:10.1099/mic.0.039834-0
Yeh HY, Chen TC, Liou KM, Hsu HT, Chung KM, Hsu LL, Chang BY (2011) The core-independent promoter-specific interaction of primary sigma factor. Nucleic Acids Res 39(3):913–925. doi:10.1093/nar/gkq911
Zheng JT, Wang SL, Yang KQ (2007) Engineering a regulatory region of jadomycin gene cluster to improve jadomycin B production in Streptomyces venezuelae. Appl Microbiol Biotechnol 76(4):883–888. doi:10.1007/s00253-007-1064-z
Zhuo Y, Zhang W, Chen D, Gao H, Tao J, Liu M, Gou Z, Zhou X, Ye BC, Zhang Q, Zhang S, Zhang LX (2010) Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis. Proc Natl Acad Sci U S A 107(25):11250–11254. doi:10.1073/pnas.1006085107
Acknowledgments
This work was supported by Key Program of Zhejiang Provincial Natural Science Foundation of China (LZ12C01001), National Basic Research Program of China (973 Program) (No. 2012CB721005), and Specialized Research Fund for the Doctoral Program of Higher Education (No. 20120101110143). We specially thank Dr. Gerald Zvobgo for reading this paper. We sincerely thank Chris Wood for English editing and proofreading. We are grateful to Dr. Yi-Ling Du for the critical advice on the work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 504 kb)
Rights and permissions
About this article
Cite this article
Liu, SP., Yu, P., Yuan, PH. et al. Sigma factor WhiGch positively regulates natamycin production in Streptomyces chattanoogensis L10. Appl Microbiol Biotechnol 99, 2715–2726 (2015). https://doi.org/10.1007/s00253-014-6307-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6307-1